Hepatitis B Virus
1. Morphology & Genome Organization
The human Hepatitis B Virus (HBV) is the prototype member of the family Hepadnaviridae, a group of small enveloped DNA-viruses found in mammals (woodchucks (WHV), ground squirrels (GSHBV), woolly monkeys (WMHBV)) and birds (pekin ducks (DHBV), grey herons HHBV)). Hepadnaviruses are closely related, sharing similarities in virion structure, genome organisation and replication strategy. They exhibit a narrow host range (e.g. apart from humans, HBV only infects chimpanzees) and a tissue tropism restricted mainly to the liver.
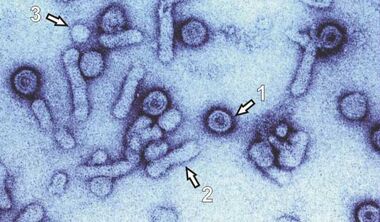
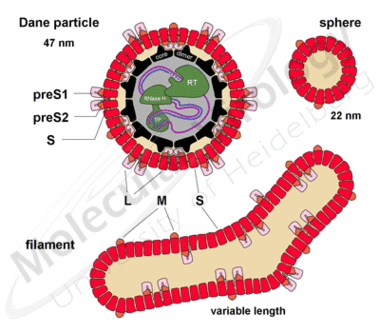
Hepadnaviruses are the smallest enveloped animal viruses with a virion diameter of 42nm for HBV (Fig.2a, 1). Interestingly, infected hepatocytes also secrete non-infectious subviral particles (SVP: filaments 2 and spheres 3) in an excess of 104 to 106 -for unknown reasons.
Fig.2a (left graphic): Electron Microscop Presentation of HBV Particles. The round 42 nm particles (1) represent infectious virions (Dane particle). The small empty spheres (3) and the filaments (3) are non infectious. The preparation was enriched in virus particles (EM picture by courtesy of H.-W. Zentgraf, Heidelberg)
Fig.2b (right graphic): Schematic representation of the three types of particles with the three envelope proteins (L, M and S) the core protein, the polymerase (RT, RNAseH and TP) and the partially double stranded DNA genome
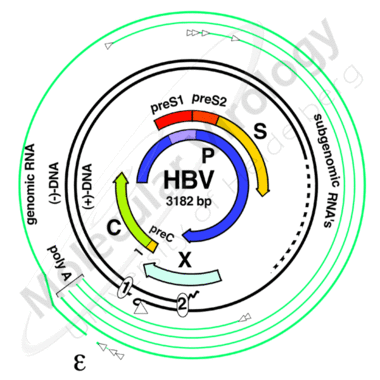
The viral genome consists of a single, partially double-stranded DNA-molecule of less than 3.200 nucleotides. It is the smallest known fully replication competent genome among animal viruses. It is remarkable for its extremely condensed organisation (Fig.3). Every single nucleotide holds protein coding function and more than half of the genome is simultaneously part of 2 overlapping open reading frames. Consequently, all of the many necessary cis-acting elements lay within a coding region (e.g. the direct repeats, the encapsidation signal, promoter- and enhancer-elements).
Fig.3:The outer lines represent the different classes of transcripts; the bold inner circles the DNA genome as present in the virion. The four major ORFs (preC/C, preS1/preS2/S, P and X) are indicated in the centre. The core (C) gene encodes the capsid protein. The P gene codes for the viral polymerase. The X gene gives rise to a protein of not completely understood function. The three envelope proteins are encoded by a single ORF with three in frame start codons.
Mammalian Hepadnaviruses utilize four major open reading frames. The core (C) gene encodes the capsid protein. The P gene codes for the viral polymerase. The X gene gives rise to a protein of not completely understood function that possesses multiple transactivating functions possibly related to HCC development.
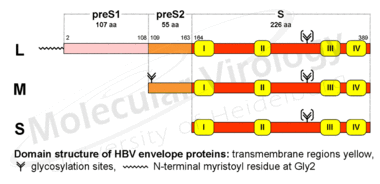
The three envelope proteins are encoded by a single ORF with three in frame start codons. Consequently, they share a common C-terminus that provides the membrane anchor and differ in length at their N-termini (Fig.4). They are designated the Large (L-), Middle (M-) and Small (S-) surface protein, respectively, with the first-mentioned playing the major role in this proposal. Compared to the S-protein (the classic HBs-Ag), the M-protein is extended to the 55 preS2- and the L-protein additionally to the 108 preS1-encoded amino acids (numbers according to subtype ayw).
Fig.4a (left graphic):The three envelope proteins of HBV (L, M and S) share a common C-terminus that provides the membrane anchor and differ in length at their N-termini (left). Compared to the S-protein (the classic HBsAg), the M-protein is extended to the 55 preS2- and the L-protein additionally to the 108 preS1-encoded amino acids.
Fig.4b (right graphic): The HBV L-protein has the unique feature of adopting at least two different topologies, one with the preS-domain located at the outside of the particle, one directed to the inside.
< Overview Page 2 (Replication) >