CLIOMMICS - Clinically-applicable, omics-based assessment of survival, side effects, and targets in multiple myeloma
Coordinator:
Prof. Dr. Hartmut Goldschmidt, Medical Clinic V, Section Multiple Myeloma, University Hospital Heidelberg
Sub project B1:
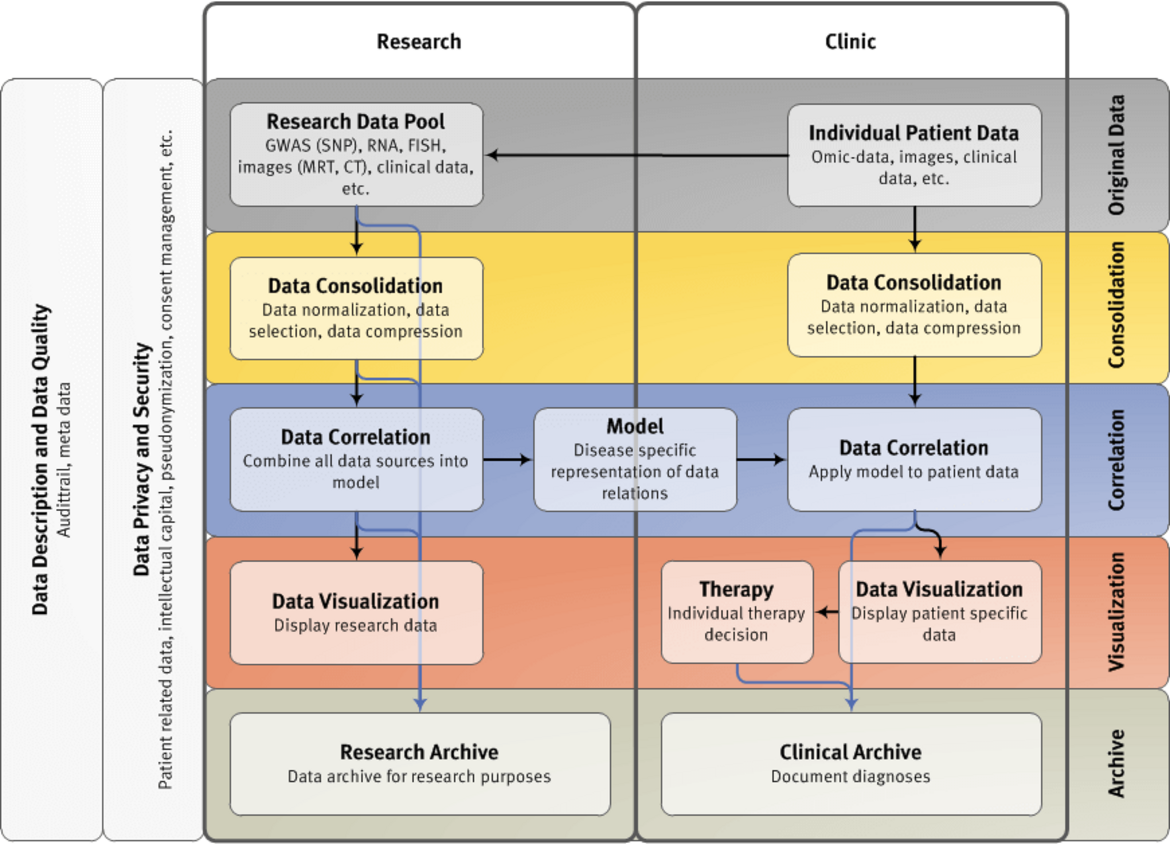
Multi-level data management and IT architecture for systems medicine for multiple myeloma
Principal investigator:
Prof. Dr. Petra Knaup-Gregori
Project partner:
Prof. Dr. Kari Hemminki, German Cancer Research Center, Heidelberg
Dr. Dirk Hose, Laboratory for myeloma research, Medical Clinic V, University Hospital Heidelberg
Prof. Dr. Annette Kopp-Schneider, German Cancer Research Center Heidelberg
Project sponsor:
e:Med-Initiative of the German ministry of education and research (BMBF), grant id: 01ZX1309A
Summary:
Multiple myeloma is a rarely curable malignant disease of clonal plasma cells which accumulate in the bone marrow. Medical challenges in multiple myeloma are threefold: i) Prognostication. Patients show different survival times ranging from a few months to 15 or more years. ii) Response to treatment. The most effective single agents (e.g. lenalidomide, bortezomib) are active in only 30% of patients. Next-generation-sequencing identified infrequent lesions but for which clinical inhibitors exist. iii) Therapy limiting side effects are frequently induced, e.g. 40% of patients treated with bortezomib develop a neuropathy. Systems-medicine challenges represent management, analysis, integration, and reporting of -omics data (transcriptomics based on RNA-sequencing, GWAS) within a timespan and in a validated way allowing clinical decisions to be drawn.
The overall research goal is the use of “omics” (GEP, RNA-sequencing, and GWAS) in clinical routine and their integration with conventional clinical (e.g. advanced imaging) and molecular (e.g. iFISH) techniques in personalizing myeloma treatment (B4); both to increase efficacy (by adding novel compounds) and simultaneously decrease the rate of side effects (by substituting compounds in patients predicted to likely suffer from their side effects). Data for prognostication focus on B3 (transcriptomics), prediction of side effects on B2 (GWAS), assessment of targets present only in a subfraction of patients on B3 (RNA-sequencing). B1 will provide storage of analysed data and programming the interface for clinical reporting (RNA-sequencing-report and CLIOMMICSreport).
Duration: 2013-2016
Project staff: M. Ganzinger, S. Kallus, C. Karmen