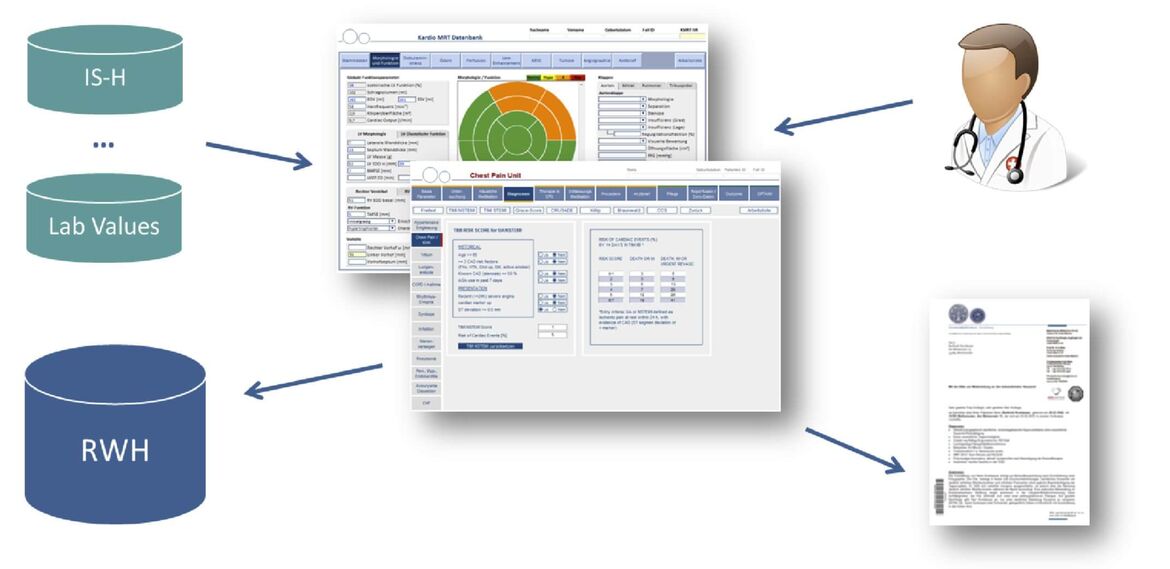
Research Data Warehouse (RWH)
To allow sophisticated clinical and translational research, clinical scientists need access to a multitude information about patients’ characteristics such as diagnoses, genetic predisposition, disease phenotype, treatment, clinical course and outcome. To reduce the amount of time spent on reading written exams for the purpose of research, the “Research Data Warehouse” (RWH) was created in 2013 with the goal of providing a platform for analyzing structured patient data already existing in the department. So far, this platform has been used to perform retrospective studies as well as to select cohorts based on a specific phenotype for genetic analyses or for the enrollment in prospective studies.
Within the Informatics for Life (www.informatics4life.org) project we develop information extraction and linkage methods to detect, retrieve, and link features such as diagnoses, medication et cetera from existing written records and integrate them with the RWH database.
Patient Reported History / Outcome
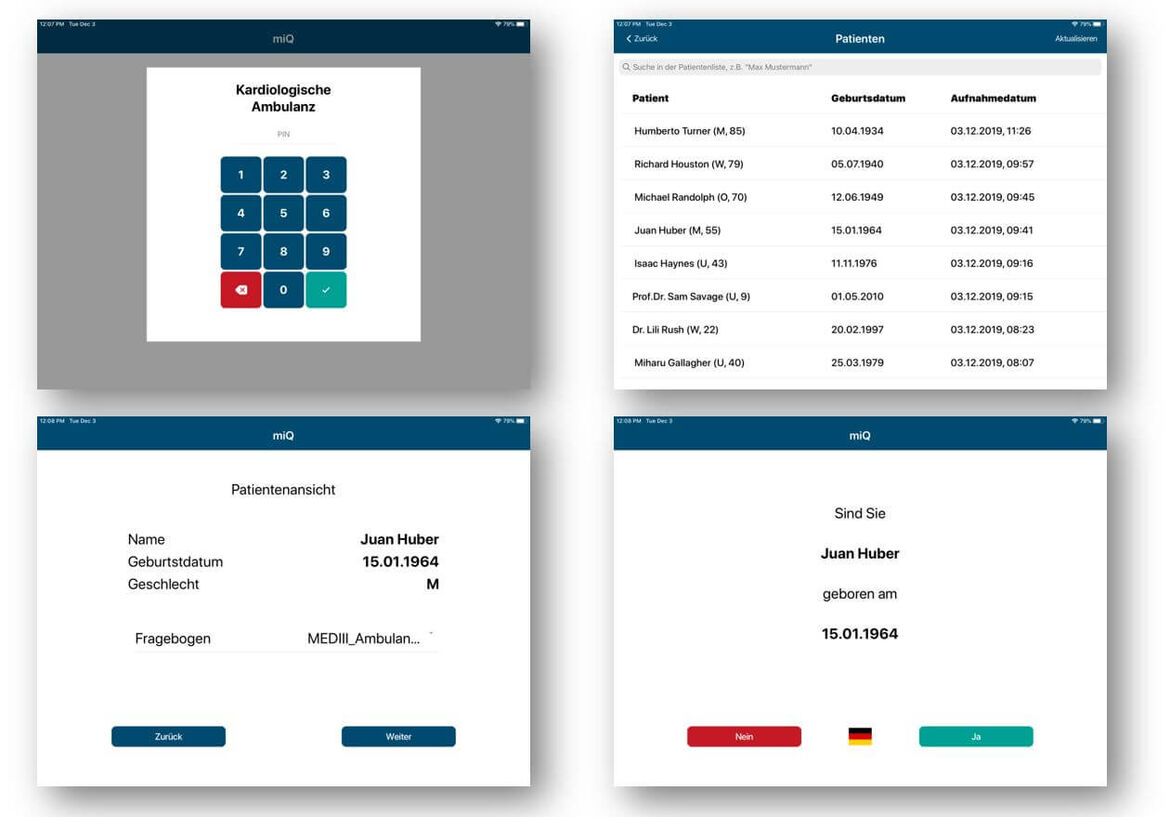
To enable a better pre- and post-clinical characterization of patients, we propose to use web-basedand mobile systems for patient reported history and outcome. This also implies motivating patientsto document their history, symptoms, risk factors, current medication, and general health status aswell as quality of life indicators after discharge from the hospital by giving them secure access totheir medical records and integrating features for scheduling appointments.
Furthermore, patientmotivation might be increased by providing information for inter-individual comparison within disease cohorts, access to disease specific patient literature as well as monitoring of relevant parameters to provide an early warning system for disease deterioration. We implement required mobile and web-based system using current standards for exchanging electronic healthcare records like IHE and HL7 FHIR.
Patient Follow-Up Supporting System
Studies evaluating treatment options rely heavily on patient outcome such as mortality and hospitalization.
To enable improved follow-up recording we establish a standardized minimal questionnaire within the cardiology department for obtaining the most relevant outcome parameters. Within this work-package we create a web-based intranet-deployed follow-up recordingsystem accessible to all cardiology personnel, enabling better data sharing between various researchgroups of the department. The proposed system integrates existing information like telephonenumbers and already retrieved outcome information as well as a workflow-engine to optimize dataacquisition and minimize redundant patient contacts.A system like this could be the basis for a centralized data acquisition unit employing study nursesto conduct telephone interviews at predefined intervals for all cardiology patients.
Information Extraction from Medical Texts
Structured medical health records of the past ten years from existing documentation systems arereadily available through the Research Warehouse at the Department of Cardiology. In addition,there is an impressive electronic archive of less structured patient information contained in, e.g.discharge letters and doctoral reports that encompass a body of more than 200.000 documents.Together, both resources are extremely valuable and will be cross-referenced with one another insupport of analysis and exploration of medical information. Basis for such cross-referencing andexploration are information networks that are constructed from entities and concepts extractedfrom medical and clinical texts. Such networks will allow to correlate and explore concepts in different contexts (over time) and thus uncover latent relationships and dependencies in diverse collectionsof text data. This work package supports our overall goal to advance individual patient care by the joint analysis of structured health records and discharge letters and will focus on the extraction of diagnoses, medication and ECG findings.
RWH User Interface for Clinical Scientists
Project-specific RWH data analysis, composition and extraction are currently performed by RWHadministrators. To enable direct data access by clinical scientists without informatics background,we propose to implement an intranet-deployed Graphical User Interface (GUI). This interface willallow clinical users to directly initiate RWH data requests and perform queries to identify patientcohorts. Identifying patient cohorts with the new GUI will use the existing RWH data model. Basedon Jim Gray’s 20 question approach we will tailor the GUI to optimize human computer interactionand use index-structures to execute cohort queries in real-time or close to real time.
The available data will be organized using a domain-specific ontology, thus providing a well-structured meta-datarepository for parameter discovery and selection. In addition, the application will replace the currently used paper-based project approval by the RWH board. In summary, the GUI will strive tosupport the entire RWH data usage process from initiating the data request to the final data transfer.