Hepatitis C Virus
1. Genome Organization & Viral Protein Expression
HCV has been classified as a member of the genus Hepacivirus that together with the genera Pestivirus and Flavivirus belongs to the family Flaviviridae. These viruses have in common a single stranded RNA genome of positive polarity that is composed of a 5' non-translated region (NTR), a long open reading frame encoding a polyprotein and a 3' NTR. In case of HCV the ORF encodes an about 3,000 amino acids long polyprotein that is translated via an internal ribosome entry site (IRES) residing in the 5´ NTR. The polyprotein is cleaved by host cell enzymes and two viral proteases into at least 10 different products. An eleventh protein -designated ARF-P or F-protein or core+1- is generated by internal translation initiation or ribosomal frame shift.
Specific functions have been ascribed to most HCV proteins. Core (C) is an RNA binding protein forming the nucleocapsid. E1 and E2 are highly glycosylated type 1 transmembrane proteins forming stable heterodimers. They are embedded into the lipid envelope surrounding the viral nucleocapsid. P7 is a small hydrophobic peptide that may be a virioporin. Most of the nonstructural proteins NS2 - NS5B are required for multiplication of the viral RNA. NS2 and the amino terminal domain of NS3 constitute the NS2/3 autoprotease responsible for cleavage at the NS2/3 junction. NS3 is a bifunctional molecule carrying three different enzymatic activities. The aminoterminal domain forms a serine-type protease responsible for processing at all sites carboxy terminal of NS3 and the carboxy terminal domain carries NTPase/ helicase activities.
Apart from the NS proteins, cis acting RNA elements in the 5' and 3' NTRs are required for RNA replication. Moreover, a stem-loop structure in the 3' terminal coding region of NS5B is essential for RNA multiplication. Genetic studies suggest that this RNA element is engaged in base pairing with a hairpin structure in the 3' NTR forming a 'kissing-loop' interaction that is essential for RNA replication.
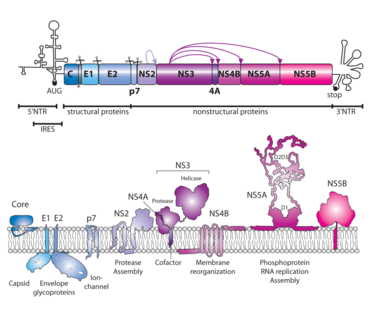
A schematic representation of the HCV genome with the 5' and 3' NTRs is shown on the top. The two viral proteases involved in processing of the polyprotein are indicated by arrows with arrow heads pointing to the cleavage sites. Scissors indicate cleavage sites of the host cell signal peptidase. The membrane topologies of cleaved viral proteins are given on the bottom, along with functions ascribed to the different proteins. IRES, internal ribosome entry site. After Neufeldt et al., Nat Rev Microbiol 2018.
< Overview Page 2 (Replication Scheme) >