Hepatitis C Virus
2. Replication Scheme
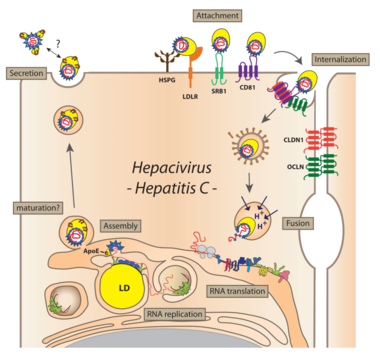
The primary host cell is the hepatocyte but replication has also been described for peripheral blood mononuclear cells (PBMCs) as well as several B and T cell lines. Upon binding of HCV to the host cell the viral RNA is released from the nucleocapsid into the cytoplasm presumably in a pH-dependent manner. Directed by the IRES in the 5' NTR, the viral RNA genome is translated and the resulting polyprotein is proteolytically cleaved into individual proteins. Most, if not all, of these proteins form a higher-order multi-protein complex tightly associated with intracellular membranes that are rearranged to form double-membrane vesicles (DMVs). Within these DMVs the positive strand RNA genome is copied into a negative strand RNA intermediate that in turn serves as a template for synthesis of excess amounts of positive strand progeny. These positive strands may be used either for synthesis of new negative strands, for protein synthesis or for encapsidation into virus particles. The fact that the E-proteins are not transported beyond the cis-Golgi suggests that viral nucleocapsids acquire their envelope by budding into the lumen of intracellular membrane compartments. In this case the virus may be exported via the constitutive secretory pathway.
Virus particles bind to the host cell via SRBI and CD81, followed by engagement with the tight junction proteins Claudin-1 and Occludin. Bound particles are internalized by receptor-mediated endocytosis. After the viral genome is liberated from the nucleocapsid (uncoating) and translated at the rough ER, NS4B and NS5A (in conjunction with other viral and cellular factors) induce the formation of DMVs accumulating in the cytoplasm and forming a structure originally called the membranous web. These membranes are supposed to serve as scaffolds for the viral replication complex. After genome amplification and structural protein production, progeny virions are assembled. The site of virus particle formation has not yet been clearly identified. It takes place most likely at intracellular membranes derived from the ER or the Golgi compartment and tightly associated with lipid droplets. Newly produced virus particles may leave the host cell by the secretory pathway.
< Page 1 (Genome Organization)