Research Group Kräusslich
Research projects in our group focus on the interaction of retroviruses with their host cells.
Projects
Research projects in our group address different aspects of retroviral biology. A long-term interest of the group is the formation and architecture of the infectious human immunodeficiency virus (HIV) particle. A major current focus in the lab is the study of entry and post-entry stages in the life cycle of HIV‑1 and murine leukemia virus and the role of innate immune responses in these processes. We combine conventional and super-resolution microscopy as well as correlative-light and electron microscopy (CLEM) to study the composition and architecture of entering subviral complexes and the interaction with cellular proteins and structures. Other projects address the role of lipids for virus assembly and cell entry, the use of CRISP/Cas based approaches for HIV‑1 elimination, and the spread of influenza virus in complex cell systems. Our main research interests are:
- Morphogenesis and structure of HIV
- Post-entry stages in retroviral replication
- Role of lipids in virus replication
Morphogenesis and Structure of HIV Particles
Assembly and budding of Human Immunodeficiency Virus (HIV) is directed by the main structural polyprotein Gag and occurs at the plasma membrane of the infected cell. The virus is initially released as immature non-infectious particle consisting of mostly uncleaved Gag polyproteins. The immature virion consists of approximately 2500 Gag molecules arranged as spherical protein shell. Particle maturation occurs after proteolytic processing of Gag at five different cleavage sites, which is mediated by the virus-encoded, virion-associated protease (PR). Maturation leads to disassembly of the immature Gag layer, followed by a second assembly stage forming the mature cone-shaped capsid that incorporates the condensed viral genome associated with replication proteins.
The assembly of immature particles requires only the viral Gag polyprotein and can be mimicked in vitro using bacterially purified Gag-derived proteins. We have developed in vitro assembly systems to analyze the molecular architecture of capsid assemblies, to study the effect of mutations on mature and immature particle formation and identify inhibitors of HIV assembly. These studies are complemented by analyses of HIV particle formation in tissue culture.
We employ structural analyses including transmission EM, cryo-EM and tomography to obtain detailed three-dimensional images of mature and immature virions, as well as viral budding sites, of wild-type and mutant HIV. These provide a better understanding of the mechanisms and protein interfaces governing assembly and maturation.
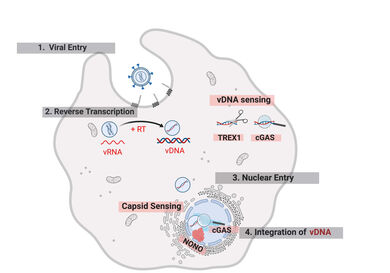
Post-Entry Stages of Retroviral Replication
Delivery of the genome-containing retroviral capsid into the cytoplasm of a newly infected cell is followed by reverse transcription of the viral RNA and transport of the resulting cDNA to the host cell DNA, where the genetic information of the virus is integrated. These early post-entry replication steps occur within poorly charaterized subviral complexes termed reverse-transcription and pre-integration complexes (RTC/PIC). Increasing evidence indicates that the viral capsid plays a central role throughout this replication phase.
A detailed understanding of post-entry is hampered by the fact that reverse transcription, intracellular transport and capsid uncoating are tightly intertwined. Furthermore, numerous host cell dependency and restricition factors affect these processes in a cell type dependent manner. Finally, there are fundamental differences not only between different types of host cells, but also between different retroviruses. While many retroviruses (e.g. moloney leukemia virus, MLV) depend on cell division to gain access to host cell genome, lentiviruses, e.g. HIV-1, can enter the intact nucleus through nuclear pores.
Our work on retroviral post-entry comprises a number of different projects, in which we
- characterize the composition and structure of retroviral replication complexes in different cells and at different intracellular localizations,
- investigate the function of host cell dependency (e.g. CPSF6, Nup153) and restriction factors that interact with the incoming viral capsid and of cellular sensors (e.g. cGAS, TREX1) that detect the viral cDNA,
- assess the role of capsid stability in post-entry processes
- and compare replication pathways and cellular immune responses in HIV-1 and MLV infected cells.
For this, we combine advanced labeling technologies (e.g. EdU-click labeling of viral DNA, 3D FISH ANCHOR visualization) with state-of-the-art microscopy (STED nanoscopy, correlative light and electron microscopy, (cryo)electron tomography, live cell imaging), molecular biological (e.g. digital droplet PCR), virological and biochemical approaches. In these projects, we collaborate with different groups within the Collaborative Research Center SFB 1129 and the Priority Program SPP 1923, funded by the German Research Foundation (DFG).
Role of Lipids in HIV Replication
HIV is an enveloped virus. Its protein capsid is surrounded by a lipid envelope, which is derived from the host cell membrane in the viral budding process. During infection of new host cells, this lipid envelope fuses to the cellular plasma membrane. Thus, lipids are involved in virus entry and egress, and they may play functional roles in viral protein composition, particle stability and infectivity. However, unlike viral proteins, viral lipids and their functional role a poorly understood.
In collaboration with Britta Brügger we have determined the lipidome of HIV using mass spectroscopy and biochemical approaches. We are currently analyzing the influence of host cells and viral proteins on the lipid compostion of the envelope and study the influence of lipid composition on the properties of the virus.
In order to study the lipid and protein organization at HIV-1 assembly sites, we have developed a system that allows us to recruit the viral structural protein Gag to the plasma membrane at a defined time point. Using clickable lipid analogs, STED and MINFLUX nanoscopy we investigate the (re)distribution of distinct lipids during the viral assembly process.
Selected Publications
Complete Publication List (PubMed)
Müller TG, Klaus S, Zila V, Lucic B, Penzo C, Nopper SL, Golani G, Anders-Össwein M, Sonntag-Buck V, Heuser AM, Schwarz US, Laketa V, Lusic M, Müller B, Kräusslich HG. (2025) Lenacapavir-induced capsid damage uncovers HIV-1 genomes emanating from nuclear speckles. EMBO J. doi: 10.1038/s44318-025-00652-5.
- Hacke M, Kaplan C, Laketa V, Heilemann M, Müller B, Kräusslich HG (2025) Multiplex 3D Nanoscopy Resolves Nanoarchitecture of Human Immunodeficency Virus. ACS Nano doi.org/10.1021/acsnano.5c12222
- Stacey JCV, Hrebík D, Nand E, Dyavari Shetty S, Qu K, Boicu M, Anders-Össwein M, Uchil PD, Dick RA, Mothes W, Kräusslich HG, Müller B, Briggs JAG. (2025). The conserved HIV-1 spacer peptide 2 triggers matrix lattice maturation. Nature. doi.org/10.1038/s41586-025-08624-9
- Kreysing JP, Heidari M, Zila V, Cruz-León S, Obarska-Kosinska, A, Laketa V, Rohleder L, Welsch S,
Köfinger J, Turoňová B, Hummer G, Kräusslich HG, Beck M (2024) Passage of the HIV-1 capsid cracks the nuclear pore. Cell. doi: 10.1016/j.cell.2024.12.008
Peter RS Nieters A Göpel S Merle U Steinacker JM Deibert P Friedmann-Bette B Nieß A Müller B Schilling C Erz G Giesen R Götz V Keller K Maier P Matits L Parthé S Rehm M Schellenberg J Schempf U Zhu M Kräusslich HG Rothenbacher D Kern WV EPILOC Phase 2 Study GroupPeter RS Nieters A Göpel S Merle U Steinacker JM Deibert P Friedmann-Bette B Nieß A Müller B Schilling C Erz G Giesen R Götz V Keller K Maier P Matits L Parthé S Rehm M Schellenberg J Schempf U Zhu M Kräusslich HG Rothenbacher D Kern WV EPILOC Phase 2 Study Group. (2025) Persistent symptoms and clinical findings in adults with post-acute sequelae of COVID-19/post-COVID-19 syndrome in the second year after acute infection: A population-based, nested case-control study. PLoS Medicine. 22(1): e1004511. doi: 10.1371/journal.pmed.1004511
- Peter R, Nieters A, Kräusslich HG, Brockmann SO, Göpel S, Kindle G, Merle U, Steinacker J, Rothenbacher D, Kern W (2022) Post-acute sequelae of covid-19 six to 12 months after infection: population based study. BMJ. doi: https://doi.org/10.1136/bmj-2022-071050
- Müller, TG, Zila V, Müller B, Kräusslich HG (2022) Nuclear Capsid Uncoating and Reverse Transcription of HIV-1. Annual Reviews of Virology 9:20.1–20.24. doi.org/10.1146/annurev-virology-020922-110929
- Zila V, Müller TG, Müller B, Kräusslich HG (2021) HIV-1 capsid is the key orchestrator of early viral replication. PLoS Pathog 17(12):e1010109. doi: 10.1371/journal.ppat.1010109.
- Qu, K., Ke, Z., Zila, V. Anders-Össwein, M., Glass, B., Mücksch, F., Müller, R., Schultz, C., Müller, B., Kräusslich, H.G. and Briggs, J.A.G. (2021) Maturation of the matrix and viral membrane of HIV-1. Science, Vol. 373, Issue 6555, pp. 700-704. DOI: 10.1126/science.abe6821
- Müller TG, Zila V, Peters K, Schifferdecker S, Stanic M, Lucic B, Laketa V, Lusic M, Müller B, Kräusslich HG. HIV-1 uncoating by release of viral cDNA from capsid-like structures in the nucleus of infected cells. Elife. 2021 Apr 27;10:e64776. doi: 10.7554/eLife.64776. Epub ahead of print. PMID: 33904396.
- Zila V, Margiotta E, Turoňová B, Müller TG, Zimmerli CE, Mattei S, Allegretti M, Börner K, Rada J, Müller B, Lusic M, Kräusslich HG, Beck M. (2021). Cone-shaped HIV-1 capsids are transported through intact nuclear pores. Cell. S0092-8674(21)00068-4. doi: 10.1016/j.cell.2021.01.025
- Tönshoff B, Müller B, Elling R, Renk H, Meissner P, Hengel H, Garbade SF, Kieser M, Jeltsch K, Grulich-Henn J, Euler J, Stich M, Chobanyan-Jürgens K, Zernickel M, Janda A, Wölfle L, Stamminger T, Iftner T, Ganzenmueller T, Schmitt C, Görne T, Laketa V, Olberg S, Plaszczyca A, Cortese M, Bartenschlager R, Pape C, Remme R, Huzly D, Panning M, Weigang S, Giese S, Ciminski K, Ankerhold J, Kochs G, Schwemmle M, Handgretinger R, Niemeyer CM, Engel C, Kern WV, Hoffmann GF, Franz AR, Henneke P, Debatin KM, Kräusslich HG. (2021) Prevalence of SARS-CoV-2 Infection in Children and Their Parents in Southwest Germany.JAMA Pediatr. 2021 Jan 22. doi: 10.1001/jamapediatrics.2021.0001.
- Peukes J, Xiong X, Erlendsson S, Qu K, Wan W, Calder LJ, Schraidt O, Kummer S, Freund SMV, Kräusslich HG, Briggs JAG (2020) The native structure of the assembled matrix protein 1 of influenza A virus. Nature 587(7834):495-498
- Mücksch, F., Citir, M., Lüchtenborg, C, Glass, B., Traynor-Kaplan, A., Schultz, C., Brügger, B. and Kräusslich, H.G. (2019) Quantification of Phosphoinositides Reveals Strong Enrichment of PIP 2 in HIV-1 Compared to Producer Cell Membranes. Sci Rep 9, 17661
- Zila, V., Müller, T.G., Laketa, V., Müller, B. and Kräusslich, H.G. (2019). Analysis of CA Content and CPSF6 Dependence of Early HIV-1 Replication Complexes in SupT1-R5 Cells. MBio, doi 10.1128/mBio.02501-19.
- Bejarano, D.A., Peng, K., Laketa, V., Börner, K., Jost, K.L., Lucic, B., Glass, B., Lusic, M., Müller, B., and Kräusslich, H.G. (2019). HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the Nuclear Pore Complex. eLife, e41800.
- Mattei S., Tan A, Glass B, Müller B., Kräusslich H.G., and Briggs J.A.G. (2018) High-resolution structures of HIV-1 Gag cleavage mutants determine structural switch for virus maturation. Proc Natl Acad Sci USA. doi.org/10.1073/pnas.181123711.5
- Mücksch F, Laketa V, Müller B, Schultz C, Kräusslich HG (2017) Synchronized HIV assembly by tunable PIP2 changes reveals PIP2 requirement for stable Gag anchoring. Elife. pii: e25287. doi: 10.7554/eLife.25287.
- Mattei S, Glass B, Hagen WJ, Kräusslich HG, Briggs JA. (2016). The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354(6318):1434-1437.
- Hanne J, Göttfert F, Schimer J, Anders-Össwein M, Konvalinka J, Engelhardt J, Müller B, Hell SW, Kräusslich HG (2016). Stimulated Emission Depletion Nanoscopy Reveals Time-Course of Human Immunodeficiency Virus Proteolytic Maturation . ACS Nano, in press.
- Schur FK, Obr M, Hagen WJ, Wan W, Jakobi AJ, Kirkpatrick JM, Sachse C, Kräusslich HG, Briggs JA (2016) An atomic model of HIV-1 capsid-SP1 reveals structures regulating assembly and maturation. Science 353(6298):506-8.
- Hanne J, Zila V, Heilemann M, Müller B, Kräusslich HG.(2016) Super-resolved insights into human immunodeficiency virus biology. FEBS Lett. 590(13):1858-76.
- Konvalinka, J., Kräusslich, H.G.***, and Müller, B. (2015). Retroviral proteases and their roles in virion maturation. Virology. doi: 10.1016/j.virol.2015.03.021.
- Schimer J, Pavova M, Anders M, Pachl P, Sacha P, Cigler P, Weber J, Majer P, Rezacova P, Kräusslich HG, Müller B***, Konvlinka J***, (2015). Triggering HIV polyprotein processing inside virions by rapid photodegradation of a tight-binding photodestructable protease Inhibitor. Nature Communications 6:6461.
- Peng K, Muranyi W., Glass B, Laketa V, Yant SR, Tsai L, Cihlar T, Müller B, Kräusslich, HG. (2014) Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. eLife 2014 10.7554/eLife.04114
- Herold N, Anders-Ößwein M, Glass B, Eckhardt M, Müller B, Kräusslich HG. (2014) HIV-1 Entry in SupT1-R5, CEM-ss, and Primary CD4+ T Cells Occurs at the Plasma Membrane and Does Not Require Endocytosis. J Virol. 88:13956-70.
- Schur, F.K.M., Hagen, W.J.H., Rumlová, M., Ruml, T., Müller, B., Kräusslich, H.G., and Briggs, J.A.G. (2014) The structure of the immature HIV-1 capsid in intact . virus particles at 8.8 Å resolution. Nature. 2014 Nov 2. doi: 10.1038/nature13838.
- Izquierdo-Useros N, Lorizate M, Puertas MC, Rodriguez-Plata MT, Zangger N, Erikson E, Pino M, Erkizia I, Glass B, Clotet B, Keppler OT, Telenti A, Kräusslich HG, Martinez-Picado J. (2013) Siglec-1 is a novel dendritic cell receptor that mediates HIV-1 trans-infection through recognition of viral membrane gangliosides. PLoS Biol. 2012;10(12):e1001448.
- Muranyi, W., S. Malkusch, B. Müller, M. Heilemann, and H. G. Kräusslich (2013) Super-resolution Microscopy Reveals Specific Recruitment of HIV-1 Envelope Proteins to Viral Assembly Sites dependent on the Envelope C-Terminal Tail. PLoS Pathog, in press.
- Chojnacki J, Staudt T, Glass B, Bingen P, Engelhardt J, Anders M, Schneider J, B. M, Hell S, Kräusslich HG (2012) Maturation Dependent HIV-1 Surface Protein Redistribution Revealed by Fluorescence Nanoscopy. Science 2012, 338:524-528.
- Izquierdo-Useros, N., Lorizate, M., Contreras, F.X., Rodriguez-Plata, M.T., Glass, B., Erkizia, I., Prado, J.G., Casas, J., Fabrias, G., Kräusslich, H.G., et al. (2012). Sialyllactose in viral membrane gangliosides is a novel molecular recognition pattern for mature dendritic cell capture of HIV-1. PLoS biology 10, e1001315.
- Radestock, B., Morales, I., Rahman, S.A., Radau, S., Glass, B., Zahedi, R.P., Müller, B., and Kräusslich, H.G. (2012). Comprehensive Mutational Analysis Reveals p6Gag Phosphorylation to be Dispensable for HIV-1 Morphogenesis and Replication. Journal of virology.
- Geis, S., Prifert, C., Weissbrich, B., Lehners, N., Egerer, G., Eisenbach, C., Buchholz, U., Aichinger, E., Dreger, P., Neben, K., et al. (2012). Molecular Characterization of a Respiratory Syncytial Virus (RSV) Outbreak in a Hematology Unit, Heidelberg, Germany. Journal of clinical microbiology.
- Sundquist, W.I., and Krausslich, H.G. (2011). HIV-1 Assembly, Budding, and Maturation. Cold Spring Harbor perspectives in medicine 2, a006924.
- Lorizate, M. and Krausslich, H.G. Role of Lipids in Virus Replication. Cold Spring Harbor perspectives in biology. 2011. cshperspectives.cshlp.org/cgi/reprint/cshperspect.a004820v1.pdf
- Briggs JA, Kräusslich HG. (2011) The molecular architecture of HIV. J Mol Biol. 2011 Jul 22;410(4):491-500J Mol Biol. 410(4):491-500
- von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomlin N, Pechstein A, Chau N, Chircop M, Sakoff J, von Kries JP, Saenger W, Kräusslich HG, Shupliakov O, Robinson PJ, McCluskey A, Haucke V. (2011) Role of the clathrin terminal domain in regulating coated pit dynamics revealed by small molecule inhibition.Cell 146(3):471-84.
- Baumgartel, V., Ivanchenko, S., Dupont, A., Sergeev, M., Wiseman, P.W., Krausslich, H.G., Brauchle, C., Muller, B., and Lamb, D.C. 2011. Live-cell visualization of dynamics of HIV budding site interactions with an ESCRT component. Nature cell biology 13(4): 469-474.
- Carlson, L.A., de Marco, A., Oberwinkler, H., Habermann, A., Briggs, J.A., Krausslich, H.G., and Grunewald, K. 2010. Cryo electron tomography of native HIV-1 budding sites. PLoS pathogens 6(11): e1001173.
- de Marco, A., Muller, B., Glass, B., Riches, J.D., Krausslich, H.G., and Briggs, J.A. 2010. Structural analysis of HIV-1 maturation using cryo-electron tomography. PLoS pathogens 6(11): e1001215.
- Habermann, A., Krijnse-Locker, J., Oberwinkler, H., Eckhardt, M., Homann, S., Andrew, A., Strebel, K., and Krausslich, H.G. 2010. CD317/tetherin is enriched in the HIV-1 envelope and downregulated from the plasma membrane upon virus infection. Journal of virology 84(9): 4646-4658.
- Ivanchenko, S., Godinez, W.J., Lampe, M., Krausslich, H.G., Eils, R., Rohr, K., Brauchle, C., Muller, B., and Lamb, D.C. 2009. Dynamics of HIV-1 assembly and release. PLoS pathogens 5(11): e1000652.
- Koch, P., Lampe, M., Godinez, W.J., Muller, B., Rohr, K., Krausslich, H.G., and Lehmann, M.J. 2009. Visualizing fusion of pseudotyped HIV-1 particles in real time by live cell microscopy. Retrovirology 6: 84.
- Lorizate, M., Brugger, B., Akiyama, H., Glass, B., Muller, B., Anderluh, G., Wieland, F.T., and Krausslich, H.G. 2009. Probing HIV-1 membrane liquid order by Laurdan staining reveals producer cell-dependent differences. The Journal of biological chemistry 284(33): 22238-22247.
- Muller, B., Anders, M., Akiyama, H., Welsch, S., Glass, B., Nikovics, K., Clavel, F., Tervo, H.M., Keppler, O.T., and Krausslich, H.G. 2009. HIV-1 Gag processing intermediates trans-dominantly interfere with HIV-1 infectivity. The Journal of biological chemistry 284(43): 29692-29703.
- Pongratz, C., Yazdanpanah, B., Kashkar, H., Lehmann, M.J., Krausslich, H.G., and Kronke, M. Selection of potent non-toxic inhibitory sequences from a randomized HIV-1 specific lentiviral short hairpin RNA library. PloS one 5(10): e13172.
- Rezacova, P., Pokorna, J., Brynda, J., Kozisek, M., Cigler, P., Lepsik, M., Fanfrlik, J., Rezac, J., Grantz Saskova, K., Sieglova, I., Plesek, J., Sicha, V., Gruner, B., Oberwinkler, H., Sedlacek, J., Krausslich, H.G., Hobza, P., Kral, V., and Konvalinka, J. 2009. Design of HIV protease inhibitors based on inorganic polyhedral metallacarboranes. Journal of medicinal chemistry 52(22): 7132-7141.
- Tebit, D.M., Lobritz, M., Lalonde, M., Immonen, T., Singh, K., Sarafianos, S., Herchenroder, O., Krausslich, H.G., and Arts, E.J. Divergent evolution in reverse transcriptase (RT) of HIV-1 group O and M lineages: impact on structure, fitness, and sensitivity to nonnucleoside RT inhibitors. Journal of virology 84(19): 9817-9830.
- Tebit, D.M., Sangare, L., Tiba, F., Saydou, Y., Makamtse, A., Somlare, H., Bado, G., Kouldiaty, B.G., Zabsonre, I., Yameogo, S.L., Sathiandee, K., Drabo, J.Y., and Krausslich, H.G. 2009. Analysis of the diversity of the HIV-1 pol gene and drug resistance associated changes among drug-naive patients in Burkina Faso. Journal of medical virology 81(10): 1691-1701.
- Welsch, S., Groot, F., Krausslich, H.G., Keppler, O.T., and Sattentau, Q. 2011. Architecture and Regulation of the HIV-1 Assembly and Holding Compartment in Macrophages. Journal of virology.
- Briggs JA, Riches JD, Glass B, Bartonova V, Zanetti G, Kräusslich HG. 2009. Structure and assembly of immature HIV. Proc Natl Acad Sci U S A. 10627:11090-5.
- Dam E, Quercia R, Glass B, Descamps D, Launay O, Duval X, Kräusslich HG, Hance AJ, Clavel F; ANRS 109 Study Group. 2009. Gag mutations strongly contribute to HIV-1 resistance to protease inhibitors in highly drug-experienced patients besides compensating for fitness loss. PLoS Pathog. 2009 Mar;5(3):e1000345. Epub 2009 Mar 20
- Carlson LA, Briggs JA, Glass B, Riches JD, Simon MN, Johnson MC, Müller B, Grünewald K, Kräusslich HG (2008) Three-dimensional analysis of budding sites and released virus suggests a revised model for HIV-1 morphogenesis.Cell Host Microbe 4:592-9.
- Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Kräusslich HG. (2007) HIV-1 buds predominantly at the plasma membrane of primary human macrophages.PLoS Pathog. 3:e36.
- Tebit DM, Sangaré L, Makamtse A, Yameogo S, Somlare H, Bado G, Kouldiaty BG, Sathiandee K, Tiba F, Sanou I, Ouédraogo-Traoré R, Zoungrana L, Diallo I, Drabo JY, Kräusslich HG. 2008. HIV drug resistance pattern among HAART-exposed patients with suboptimal virological response in Ouagadougou, Burkina Faso. J Acquir Immune Defic Syndr. 49(1):17-25
- Bruegger B, Glass B, Haberkant P, Leibrecht I, Wieland FT, Kraeusslich HG. 2006. The HIV lipidome: a raft with an unusual composition. PNAS 103:2614-2646
- Briggs JA, Grünewald K, Glass B, Forster F, Kräusslich HG, Fuller SD. 2006. The Mechanism of HIV-1 core assembly: insights from three-dimensional reconstructions of authentic virions. Structure 14:15-20.
- Sticht J, Humbert M, Findlow S, Bodem J, Müller B, Dietrich U, Werner J and Kräusslich HG. 2005. A peptide inhibitor of HIV-1 assembly in vitro. Nat Struct Mol Biol. 12:671-677
- Gottwein E and Kräusslich HG. 2005. Analysis of human immunodeficiency virus type 1 Gag ubiquitination. J Virol. 79:9134-9144
- Daecke J, Fackler OT, Dittmar MT, Kräusslich HG. 2005. Involvement of clathrin-mediated endocytosis in human immunodeficiency virus type 1 entry. J Virol. 79:1581-1594
- Bohne J, Wodrich H and Kräusslich HG. 2005. Splicing of human immunodeficiency virus RNA is position-dependent suggesting sequential removal of introns from the 5' end. Nucleic Acids Res. 33:825-837.
- Briggs JA, Simon MN, Gross I, Kräusslich HG, Fuller SD, Vogt VM and Johnson MC. 2004. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 11:672-675
- Müller B, Daecke J, Fackler OT, Dittmar MT, Zentgraf H and Kräusslich HG. 2004. Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative.J Virol.78:10803-10813
- Tebit DM, Zekeng L, Kaptue L, Gürtler L, Fackler OT, Keppler OT, Herchenröder O, Kräusslich HG. 2004. Construction and characterization of an HIV-1 group O infectious molecular clone and analysis of vpr- and nef-negative derivatives.Virology. 326:329-339.
- Briggs, J. A. G., T. Wilk, R. Welker, H.-G. Kräusslich and S. D. Fuller. 2003. The structural organization of authentic HIV-1 and its core. EMBO J. 22: 1707-1715.
- Fehrmann, F., M. Jung, R. Zimmermann and H.-G. Kräusslich. 2003. Transport of the Intracisternal A-Type Particle Gag Polyprotein to the Endoplasmic Reticulum is Mediated by Signal Recognition Particle. J. Virol. 77: 6293-6304.
- Gottwein, E., J. Bodem, B. Müller, A. Schmechel, H. Zentgraf and H.-G. Kräusslich. 2003. The Mason-Pfizer Monkey Virus PPPY and PSAP Motifs Both Contribute to Virus Release. J. Virol. 77: 9474-9485.
- von Schwedler, U., M. Stuchell, B. Müller, D. Ward, H.-Y. Chung, E. Morita, H. E. Wang, T. Davis, G.-P. He, D. M. Cimbora, A. Scott, H.-G. Kräusslich, J. Kaplan, S. G. Morham and W. I. Sundquist. 2003. The Protein Network of HIV Budding. Cell 114: 701-713.
- Weber, J., J. R. Mesters, M. Lepšík, J. Prejdová, M. Švec, J. Šponarová, P. Mlcochová, K. Skalicka, K. Stríšovský, T. Uhlíková, M. Soucek, L. Machala, M. Stanková, J. Vondrášek, T. Klimkait, H.-G. Kräusslich, R. Hilgenfeld, and J. Konvalinka. 2002. Unusual binding mode of an HIV-1 protease inhibitor explains its potency against multi-drug-resistant virus strains. J. Mol. Biol. 324: 739-754.
- Müller, B., T. Patschinsky, and H.-G. Kräusslich. 2002. The late domain containing protein p6 is the predominant phosphoprotein of HIV-1 particles. J. Virol. 76: 1015-1024.
- Wodrich, H., J. Bohne, E. Gumz, R. Welker, and H.-G. Kräusslich. 2001. A new RNA-element located in the coding region of a murine endogenous retrovirus can functionally replace the Rev/Rev responsive element system in human immunodeficiency virus type 1 Gag expression. J. Virol. 75:
Group Members
- Prof. Dr. med. Dr. h.c. Hans-Georg Kräusslich, Principal Investigator
Hans-Georg.Kraeusslich(at)med.uni-heidelberg.de
CIID Room 308, Phone: +49 6221 56-5001
- Dr. Samara Martín Alonso, postdoc
Samara.MartinAlonso(at)med.uni-heidelberg.de
CIID Room 304, Phone: +49 6221 56-1323
- Yu-Hsun Chen, PhD Student
- Bärbel Glass, Admin & Finance Assistant
Baerbel.Glass(at)med.uni-heidelberg.de
CIID Room 307, Phone: +49 6221 56-5701
Dr. med. Moritz Hacke, MD/PhD student
moritz.hacke(at)med.uni-heidelberg.de
CIID Room 302, Phone: +49 6221 56-1323- Anke-Mareil Heuser, Technician
Anke-Mareil.Heuser(at)med.uni-heidelberg.de
CIID, Phone: +49 6221 56-1323
- Dr. Laurence Jost, Scientific Coordinator
Laurence.Jost(at)med.uni-heidelberg.de
INF324 Room 323, Phone: +49 6221 56-7645
- Anezka Kramna, PhD Student
Anezka.Kramna(at)med.uni-heidelberg.de
CIID Room 304, Phone: +49 6221 56-1325
- Dr. Severina Klaus, research associate (IDIP)
e-mail
CIID Room 029, Phone: +49 6221 56-34410
- Dr. Vibor Laketa, IDIP Platform Coordinator
e-mail
CIID Room 029, Phone: +49 6221 56-34410
- Lara Rohleder, PhD Student
e-mail
CIID Room 304, Phone: +49 6221 56-1323
- Anja Schumacher, Secretary
Anja.Schumacher(at)med.uni-heidelberg.de
INF 324, 4th Floor, Phone: +49 6221 56-5002
- Jian Song, PhD Student
- Vera Sonntag-Buck, Technician
Vera.Sonntag-Buck(at)med.uni-heidelberg.de
CIID, Phone: +49 6221 56-5004
Alumni
Former Lab Members
- Yvonne Affram, PhD Student
- Dr. Hisashi Akiyama, Postdoc
- Anne Assmus, Diploma Student
- Vanda Bartonova, PhD Student
- Dr. David Bejarano, PhD Student and Postdoc
- Daniel Beyer, PhD Student
- Dr. Jochen Bodem, Postdoc
- Dr. Kathleen Boerner, scientific associate
- Jens Bohne, PhD Student
- Lorenzo Bulli, PhD Student
- Sarah Bunten, PhD Student
- Robin Burk, PhD Student and Postdoc
- Luis Castillo, PhD Student
- Dr. Jakub Chojnacki, Postdoc
- Ellen Collenberg, MD Student
- Jessica Daecke, PhD Student
- Laura Dietz, Master Student
- Christine Engeland, MD Student
- Eva Gottwein, PhD Student
- Anja Habermann, Technician
- Isabel Hagmann, Technician
- Janina Hanne, PhD Student
- Johannes Hermle, PhD Student
- Nikoas Herold, MD Student
- Volker Herrmann, PhD Student
- Dr. Katharina Höhn, Postdoc
- Tina Holdt, Technician
- Dr. Nuria Izquierdo, Guest Scientist
- Stefanie Jäger, PhD Student
- Dr. Jessica Janus, Postdoc
- Heiko Keller, Guest Scientist
- Peter Koch, PhD Student
- Dr. Susann Kummer, Postdoc
- Juliane Kutzner, PhD Student
- Dr. Maik Lehmann, Postdoc
- Dr. Maier Lorizate, Postdoc
- Ivonne Morales, PhD Student
- Vanessa Mrosek, MD Student
- Dr. Thorsten Müller, PhD student and postdoc
- Kerstin Müller, Diploma Student
- Frauke Mücksch, PhD Student
- PD Dr. Walter Muranyi, Research Associate
- Tamara Naumoska, PhD student
- Manuela Nickl, PhD Student
- Bianca Nouvertne, PhD Student
- Heike Oberwinkler, Technician
- Martin Obr, PhD Student
- Dr. Ke Peng, Postdoc
- Dr. Annett Petrich, Postdoc
- Benjamin Radestock, PhD Student
- Dr. Sandra Reuter, Postdoc
- Dr. Djordje Salai, PhD student and postdoc
- Kanokporn Sathiandee, PhD Student
- Michaela Schanne, PhD Student
- Sarah Stauffer, Master Student
- Jana Sticht, PhD Student
- Denis Manga Tebit, PhD Student
- Fabrice Tiba, PhD Student
- Max von Au, MD Student
- Sonja Welsch, PhD Student
- Dr. Marcela Wildova, Postdoc
- Dr. Vojtech Zila, Postdoc
Group Leader

Prof. Dr. med. Dr. h.c. Kräusslich is Dean of the Medical Faculty of Heidelberg University, Chairman of the Executive Board of the German Center for Infection Research (DZIF) and Speaker of the Collaborative Research Center SFB 1129
This group is located on the 3rd floor of the Center for Integrative Infectious Disease Research (CIID) at INF 344.