Research Group Fackler
Research projects in our group focus on "Cell Biology and Immunology of HIV Pathogenesis".
Projects
Our research addresses the cell biology, immunology and pathogenesis of HIV‑1 infection with an emphasis on CD4+ T lymphocytes. One focus of our studies is on the molecular mechanisms of action by which the HIV‑1 pathogenicity factor Nef reprograms host cell vesicular transport, signal transduction and motility to optimize HIV‑1 spread in the host and to accelerate disease progression. Another important aspect of our work is on the host innate immune system in HIV infection and on viral evasion mechanisms. This includes dissecting how the intrinsic immunity factor SERINC5 impairs HIV‑1 particle infectivity and how this activity is antagonized by the viral protein Nef, but also studies to elucidate which barriers prevent productive HIV‑1 infection of resting CD4+ T lymphocytes. These HIV-related studies involve the development of complex 3D culture systems for studying the relationship between host cell motility and HIV‑1 spread in tissue. Finally, we are also interested in the cell biology of CD4 T cell activation and differentiation. In this context, we particularly focus on the newly identified role of nuclear actin filament formation for CD4 T cell help.
HIV-1 Pathogenicity Factor Nef
Molecular Mechanisms
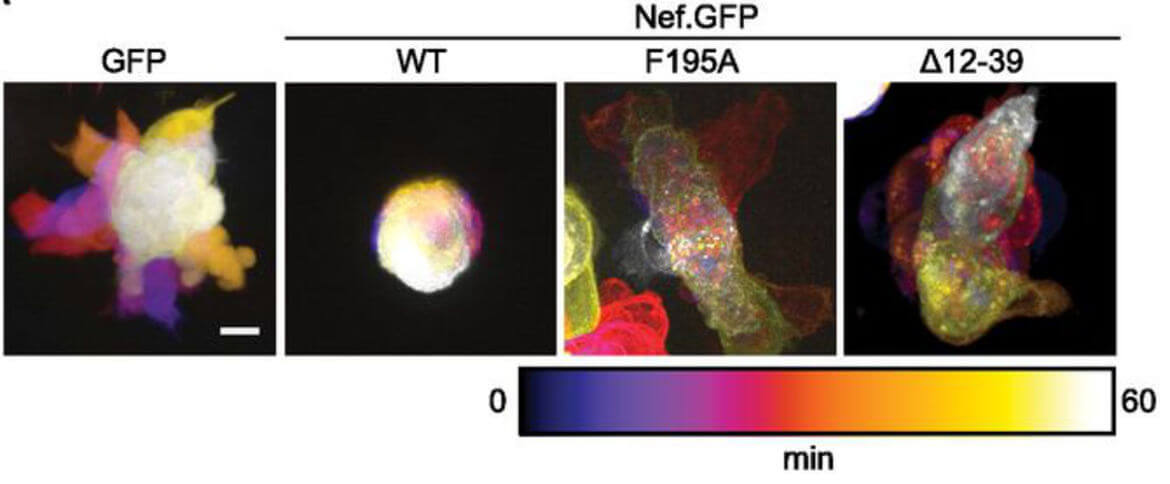
The Nef protein of HIV and SIV is critical for high virus load and full pathogenicity in the infected host. This pathogenic potential is underscored by a transgenic mouse model in which the expression of Nef alone causes an AIDS-like disease. This activity renders Nef an attractive target for antiviral strategies to complement existing treatment. Nef does not possess detectable enzymatic activity and mediates all its functions via interactions with host cell proteins. While a multitude of activities have been ascribed to Nef, the biological properties and protein-interactions that govern its pathogenic potential have still not been unambiguously identified. Nef's activities can be divided into immune evasion functions and effects that directly boost viral spread in the absence of host immune attack. Both these general Nef activities are achieved by manipulation of host cell signal transduction and intracellular transport processes. Our work focusses on visualizing and dissecting the independent molecular mechanisms employed by which Nef disrupts host cell vesicular transport, actin remodeling, and motility. This increasingly involves the use of complex primary cell culture models and in vivo analyses in mice.
Innate Sensing and Restriction of HIV-1
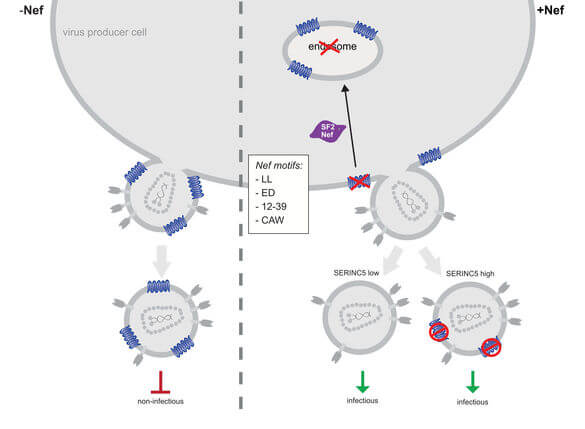
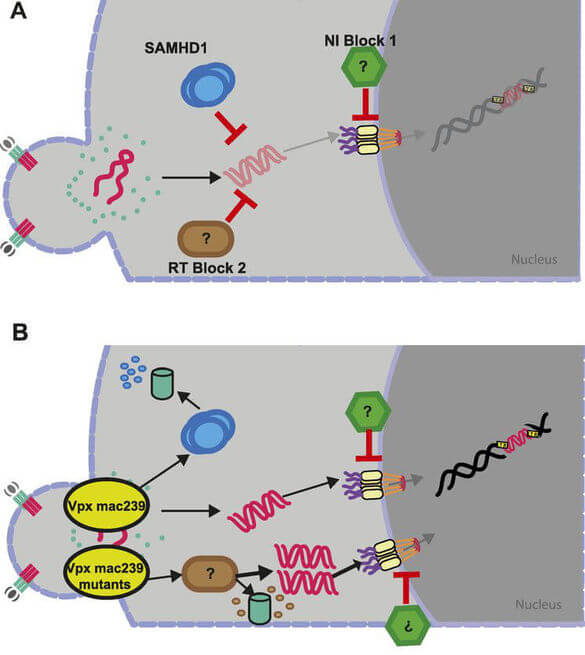
The innate immune system is increasingly recognized as a major arm of immunity against efficient HIV‑1 replication that poses cell-intrinsic barriers to virus replication (restriction factors) and elicits antiviral signaling upon recognition of replication intermediates (by sensors). Our current work focusses on two aspects of innate immune defenses against HIV.
Host Cell Restriction by SERINC Proteins
SERINC3 and 5 were recently identified as host cell restriction factors that potently suppress the infectivity of HIV‑1 virions. This antiviral activity can be counteracted by the HIV‑1 protein Nef. We are studying the mechanisms underlying the antiviral action of SERINC proteins as well as its antagonism by Nef.
HIV infection of resting CD4+ T lymphocytes
HIV‑1 infects CD4+ T lymphocytes and cells of the monocytes/macrophage lineage. In the case of primary human CD4+ T lymphocytes, the permissivity to HIV‑1 infection ex vivo depends on the cellular activation state: while activated, proliferating CD4+ T lymphocytes are highly susceptible to infection and support efficient virus replication, resting CD4+ T cells are largely non-permissive for HIV‑1 replication but can be infected in vivo and serve as a latent viral reservoir. Since physiologically the vast majority of all CD4+ T lymphocytes are in a resting state, this phenomenon provides one explanation as to why the frequency of productively infected CD4+ T lymphocytes in AIDS patients is very low. In collaboration with Prof. Dr. Oliver T. Keppler (LMU Munich) we identified the deoxynucleoside triphosphate (dNTP) triphosphohydrolase SAMHD1 (sterile alpha motif (SAM) and histidine/aspartic acid (HD) domain-containing protein 1) as an essential host cell factor for this restriction that can be overcome by the Vpx protein encoded by HIV‑2 and SIV and defined additional resting T cell-specific blocks at the level of reverse transcription and nuclear import. Our current efforts focus on defining the nature of these additional blocks to HIV‑1 replication in resting CD4+ T cells.
HIV‑1 Spread in Complex Cell Systems
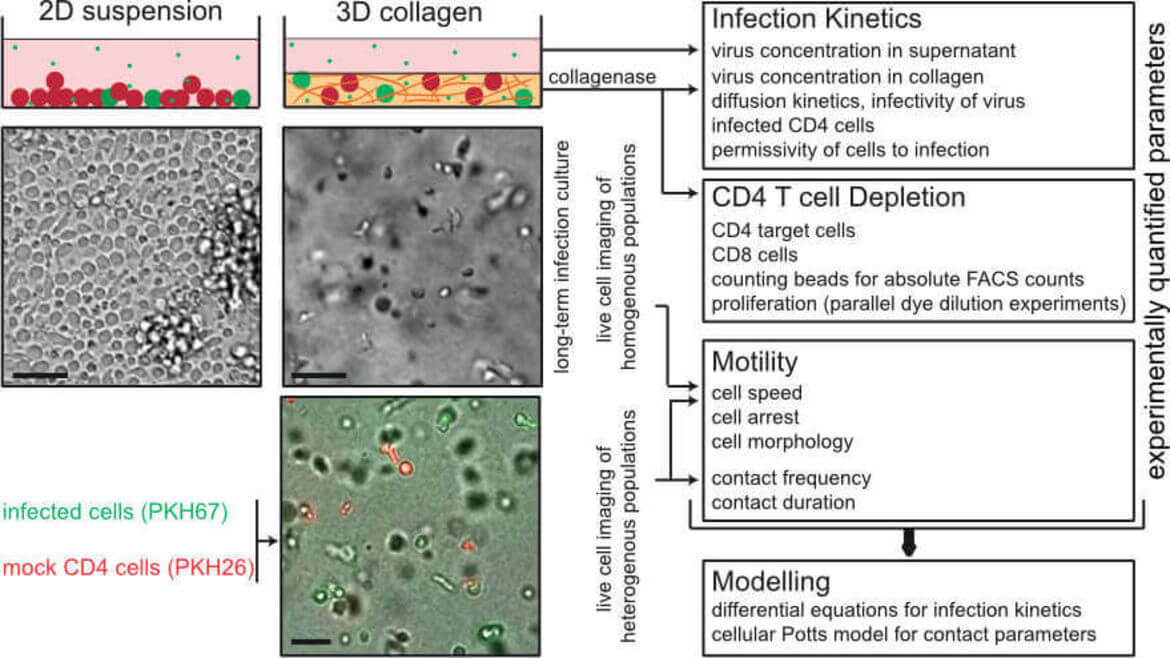
Studies on the mechanisms governing HIV‑1 spread in infected individuals are hampered by the fact that classical cell culture systems lack tissue heterogeneity and organization or are largely refractory to experimental variation of important parameters. To this end we engineered 3D collagen matrices as culture system to study HIV‑1 spread between CD4 T lymphocytes. Integration of population-based and single cell mathematical models revealed that HIV‑1 spread in 3D is predominantly mediated by cell-cell transmission and that 3D environments pose a potent restriction to cell-free infection. In turn, the 3D environment induces alterations to architecture and duration of cell-cell contacts that facilitate cell-associated HIV transfer. In our ongoing studies we focus on dissecting the relationship between cell motility and HIV spread and on increasing the complexity of this 3D ex vivo culture system.
Actin Dynamics / Nuclear Actin
Cellular Mechanisms
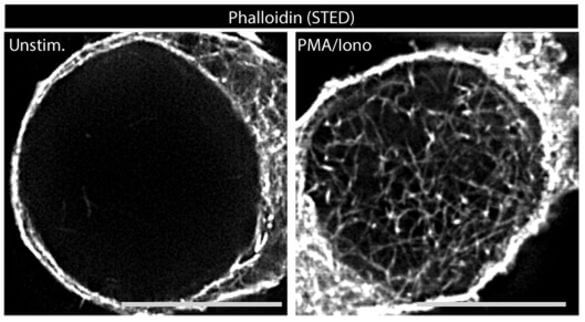
The astonishing dynamics of the actin cytoskeleton provides the basis for fundamental processes in mammalian cells such as motility, kinesis, vesicular transport and signal transduction. While mechanisms and function of actin dynamics in the cytoplasm are relatively well studied, an only recently emerging field is the role of actin dynamics in the nucleus. We recently identified the rapid and transient formation of nuclear actin filaments as novel effector function of T cell receptor stimulation in CD4 T lymphocytes. Mediated by calcium transients in the nucleus and nuclear pools of the actin polymerization machinery Arp2/3 complex, the presence of nuclear filaments facilitates expression of a selected set of genes required for CD4 T helper function. Our current efforts aim at characterizing the mechanisms governing these nucleus-specific actin polymerization event as well as at dissecting the relationship between nuclear actin dynamics and gene expression.
Selected Publications
Complete Publication List (PubMed)
- Albanese, M., Chen, H. R., Gapp, M., Muenchhoff, M., Yang, H. H., Peterhoff, D., Hoffmann, K., Xiao, Q., Ruhle, A., Ambiel, I., Schneider, S., Mejías-Pérez, E., Stern, M., Wratil, P. R., Hofmann, K., Amann, L., Jocham, L., Fuchs, T., Ulivi, A. F., Besson-Girard, S., … Fackler, O. T. and Keppler, O. T. (2024). Receptor transfer between immune cells by autoantibody-enhanced, CD32-driven trogocytosis is hijacked by HIV‑1 to infect resting CD4 T cells. Cell reports. Medicine, 5(4), 101483. https://doi.org/10.1016/j.xcrm.2024.101483
- Morath, K., Sadhu, L., Dyckhoff, G., Gapp, M., Keppler, O. T., and Fackler, O. T. (2024). Activation-neutral gene editing of tonsillar CD4 T cells for functional studies in human ex vivo tonsil cultures. Cell reports methods, 4(1), 100685. https://doi.org/10.1016/j.crmeth.2023.100685
- Sadhu, L., Tsopoulidis, N., Hasanuzzaman, M., Laketa, V., Way, M., and Fackler, O. T. (2023). ARPC5 isoforms and their regulation by calcium-calmodulin-N-WASP drive distinct Arp2/3‑dependent actin remodeling events in CD4 T cells. eLife, 12, e82450. https://doi.org/10.7554/eLife.82450
- Ananth S, Ambiel I, Schifferdecker S, Müller TG, Wratil PR, Mejias-Perez E, Kräusslich HG, Müller B, Keppler OT, Fackler OT. (2024). Spatial resolution of HIV-1 post-entry steps in resting CD4 T cells. Cell Rep. 2024 Mar 12;43(3):113941. doi: 10.1016/j.celrep.2024.113941
- Gallucci L, Abele T, Fronza R, Stolp B, Laketa V, Sid Ahmed S, Flemming A, Müller B, Göpfrich K, Fackler OT. (2023) Tissue-like environments shape functional interactions of HIV-1 with immature dendritic cells. EMBO Rep. Jun 5;24(6):e56818. doi: 10.15252/embr.202356818.
- Stolp, B., Stern, M., Ambiel, I., Hofmann, K., Morath, K., Gallucci, L., Cortese, M., Bartenschlager, R., Ruggieri, A., Graw, F., Rudelius, M., Keppler, O.T., Fackler, O.T. (2022). SARS-CoV‑2 variants of concern display enhanced intrinsic pathogenic properties and expanded organ tropism in mouse models. Cell Reports 38(7):110387.
- Albanese, M., Ruhle, A., Mittermaier, J., Mejias-Perez, E., Gapp, M., Linder, A., Schmacke, N.A., Hofmann, K., Hennrich, A.A., Levy, D.N., Humpe, A., Conzelmann, K.-K., Hornung, V., Fackler, O.T., Keppler, O.T. (2022). Rapid, efficient and activation-neutral gene editing of polyclonal primary human resting CD4+ T cells allows complex functional analyses. Nature Methods 19(1):81–89.
- Reif, T., Dyckhoff, G., Hohenberger, R., Kolbe, C.-C., Gruell, H., Klein, F., Latz, E., Stolp, B., Fackler, O.T. (2021). Contact-dependent inhibition of HIV‑1 replication in ex vivo human tonsil cultures by polymorphonuclear neutrophils. Cell Report Medicine 2(6):100317.
- Pierini, V., Gallucci, L., Stürzel, C., Kirchhoff, F., and Fackler, O.T. (2021). SERINC5 can Enhance Pro-inflammatory Cytokine Production by Primary Human Myeloid Cells in Response to Challenge with HIV‑1 Particles. J. Virol., published online Jan 17, 2021, doi.org/10.1128/JVI.02372–20.
- Kaw, S., Ananth, S., Tsopoulidis, N., Morath, K., Coban, B.M., Hohenberger, R., Bulut, O.C., Klein, F., Stolp, B., and Fackler, O.T. (2020). HIV-1 infection of CD4 T cells impairs antigen-specific B cell function. Embo J 39, e105594.
- Sid Ahmed, S., Bundgaard, N., Graw, F. and Fackler, O.T. (2020). Environmental Restrictions: A New Concept Governing HIV‑1 Spread Emerging from Integrated Experimental-Computational Analysis of Tissue-Like 3D Cultures. Cells 9: E1112.
- Stolp, B., Thelen, F., Ficht, X., Altenburger, L.M., Ruef, N., Inavalli, V., Germann, P., Page, N., Moalli, F., Raimondi, A., Keyser, K.A., Jafari, S.M.S., Barone, F., Dettmer, M.S., Merkler, D., Iannacone, M., Sharpe, J., Schlapbach, C., Fackler, O.T., Naegerl, U.V. and Stein, J.V. (2020). Salivary gland macrophages and tissue-resident CD8(+) T cells cooperate for homeostatic organ surveillance. Sci Immunol 5.
- Ananth, S., Morath, K., Trautz, B., Tibroni, N., Shytaj, I.A., Obermaier, B., Stolp, B., Lusic, M. and Fackler, O.T. (2020). Multifunctional Roles of the N‑Terminal Region of HIV‑1SF2Nef Are Mediated by Three Independent Protein Interaction Sites. J. Virol., ePub ahead of print: doi.org/10.1128/JVI.01398-19.
- Imle, A., Kumberger, P., Schnellbächer, N.D., Fehr, J., Carrillo-Bustamante, P. Ales, J., Schmidt, P., Ritter, C., Godinez, W.J., Müller, B., Rohr, K., Hamprecht, F.A., Schwarz, U.S., Graw, F., Fackler, O.T. (2019) Experimental and computational analyses reveal that environmental restrictions shape HIV‑1 spread in 3D cultures. Nat Commun, 10:2144.
- Tsopoulidis, N., Kaw, S., Laketa, V., Kutscheidt, S., Baarlink, C., Stolp, B., Grosse, R., Fackler, O.T. (2019) T cell receptor-triggered nuclear actin network formation drives CD4+ T cell effector functions. Sci Immunol, 4. pii: eaav1987.
- Lamas-Murua, M., Stolp, B., Kaw, S., Thoma, J., Tsopoulidis, N., Trautz, B., Ambiel, I., Reif, T., Arora, S., Imle, A., Tibroni, N., Wu, J., Cui, G., Stein, J.V., Tanaka, M., Lyck, R. and Fackler, O.T. (2018). HIV‑1 Nef Disrupts CD4+ T Lymphocyte Polarity, Extravasation and Homing to Lymph Nodes via its Nef-Associated Kinase Complex Interface. J. Immunol., 201:2731-2743.
- Trautz, B., Wiedemann, H., Lüchtenborg, C., Pierini, V., Kranich, J., Glass, B., Kräusslich, H.G., Brocker, T., Pizzato, M., Ruggieri, A., Brügger, B. and Fackler, O.T. (2017). SERINC5 restricts HIV‑1 infectivity without altering the lipid composition and organization of viral particles. J. Biol. Chem., 292:13702-13713.
- Baldauf, H.M., Stegmann, L., Schwarz, S.M., Ambiel, I., Trotard, M., Martin, M., Burggraf, M., Lenzi, G.M., Lejk, H., Pan, X., Fregoso, O.i., Lim, E.S., Abraham, L., Nguyen, L., Rutsch, F., König., R., Kim., B., Emerman, M., Fackler, O.T. * and Keppler, O.T. * (2017). Vpx overcomes a SAMHD1-independent block to HIV reverse transcription that is specific to resting CD4 T cells. Proc. Natl. Acad. Sci. USA, 114: 2729-2734. (*corresponding authors).
- Trautz, B., Pierini, V., Wombacher, R., Stolp, B., Chase, A.J., Pizzato, M. and Fackler, O.T. (2016). The Antagonism of HIV‑1 Nef to SERINC5 Particle Infectivity Restriction Involves the Counteraction of Virion-Associated Pools of the Restriction Factor. J. Virol., 90:10915-10927.
- Galaski, J., Ahmad, F., Tibroni, N., Pujol, F.M., Müller, B., Schmidt, R.E., and Fackler, O.T. (2015). Cell Surface Downregulation of NK Cell Ligands by Patient-derived HIV‑1 VPU and Nef Alleles. J. AIDS, 72:1-10.
- Imle, A., Abraham, L., Tsopoulidis, N., Hoflack, B., Saksela, K. and Fackler, O.T. (2015). Association with PAK2 Enables Functional Interactions of Lentiviral Nef Proteins with Exocyst. mBio, 6: e01309-15.
- Haller, C., Müller, B., Fritz, J.V., Lamas-Murua, M., Stolp, B., Pujol, F., Keppler, O.T. and Fackler, O.T. (2014). HIV‑1 Nef and Vpu are Functionally Redundant Broad-Spectrum Modulators of Cell Surface Receptors Including Tetraspanins. J. Virol., 88: 14241–14257.
- Fackler, O.T. *, Murooka, T.T., Imle, A. and Mempel, T.R.*(2014) Adding new dimensions: Towards an integrative understanding of HIV‑1 spread. (* corresponding authors). Nat. Rev. Microbiol., 12:563-574.
- Kutscheidt, S., Zhu, R., Antoku, S., Luxton, G.G.W., Stagljar, I., Fackler, O.T. * and Gundersen, G. * (2014). FHOD1 interaction with nesprin-2G mediates TAN line formation and nuclear movement (* corresponding authors). Nat. Cell Biol. 16: 708-715.
- Baldauf, H‑M.+, Pan, X.+, Erikson, E., Schmidt, S., Daddacha, W., Burggraf, M., Schenkova, K., Ambiel, I., Wabnitz G., Gramberg, T., Panitz, S., Flory, E., Landau, N.R., Sertel, S., Rutsch, F., Lasitschka, F., Kim, B., König, R., Fackler, O.T.* and Keppler, O.T.* (2012). The deoxynucleoside triphosphate triphosphohydrolase SAMHD1 restricts HIV‑1 infection in resting CD4+ T cells. Nat Med, 18: 1682–1687 (* corresponding authors, + first authors).
- Pan, X., Rudolph, J.M., Abraham, L., Habermann, A., Haller, C., Krijnse-Locker, J. and Fackler, O.T. (2012) HIV‑1 Nef compensates for disorganization of the immunological synapse by inducing trans-Golgi network–associated Lck signaling. Blood, 119:786-797.
- Stolp, B., Imle, A., Coelho, F.M., Hons, M., Mendiz, R.G., Lyck, R., Stein, J.V. and Fackler, O.T. (2012). HIV‑1 Nef Interferes With T Lymphocyte Circulation Through Confined Environments in vivo. Proc. Natl. Acad. Sci. USA, 109: 18541-18546.
- Stolp, B., Raichman-Fried, M., Abraham. L., Pan, X., Giese, S.I., Hannemann, S., Goulimari, P., Raz, E., Grosse, R. and Fackler, O.T. (2009). HIV‑1 Nef interferes with host cell motility by deregulation of cofilin. Cell Host and Microbe, 6:174-186
Group Members
- Prof. Dr. Oliver Fackler, Principal Investigator
Oliver.Fackler(at)med.uni-heidelberg.de
CIID Room 203, +49 6221 56-1322
- Dr. Samy Sid Ahmed, Postdoc
Samy.SidAhmed(at)med.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Ina Ambiel, Research Asscociate
Ina.Ambiel(at)med.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Dr. Swetha Ananth, Postdoc
Swetha.Ananth(at)med.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Dr. Kathrin Bajak, Project Coordinator
Kathrin.Bajak(at)med.uni-heidelberg.de
CIID Room 201, +49 6221 56-5061
- Md Hasanuzzaman, PhD Student
Md.Hasanuzzaman(at)med.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Mahalakshmi Bhat, PhD Student
MahalakshmiUmapathi.Bhat(at)med.uni-heidelberg.de
INF 324 Room 409, +49 6221 56-5006
- Katharina Morath, PhD Student
K.Morath(at)stud.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Cristina Rodriguez, PhD Student
Cristina.Rodriguez(at)mr.mpg.de
INF 324 Room 409, +49 6221 56-5006
- Carolyn Schneider, PhD Student
CarolynPetra.Schneider(at)med.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Dr. Bettina Stolp, Postdoc
Bettina.Stolp(at)med.uni-heidelberg.de
CIID Room 201, +49 6221 56-34631
- Nadine Tibroni, Research Asscociate
Nadine.Tibroni(at)med.uni-heidelberg.de
CIID Room 223, +49 6221 56-5006
- Martin Theis Uerlich, PhD Student
martin.uerlich@dkfz-heidelberg.de
CIID Room 223, +49 6221 56-5006
Alumni
Former Lab Members
- Libin Abraham (Postdoc, University of British Colombia, Vancouver)
- Sakshi Arora (Postdoc, University of Pennsylvania)
- Amanda Chase (Grant Writer and Project Manager, Stanford University)
- Jingyan Chen
- Wioleta Chmielewicz
- Bahar Melis Coban
- Leon Daniau
- Joelle Fritz (Project Manager, ParkinsonNet Luxembourg)
- Johanna Galaski (Postdoc, Jerusalem University)
- Judith Gasteier (Technical Leader, Abbot)
- Miriam Geist (Sanofi, Frankfurt)
- Simone Giese
- Claudia Haller (Research Consultant, Heidelberg Faculty of Medicine)
- Sebastian Hannemann (Quality Management Global, West Pharmaceutical Services, Eschweiler)
- Stefanie Homann (Postdoc, University of California, San Diego)
- Andrea Imle (Scientist, BioNTech)
- Sheetal Kaw (Project Head, Ablynx/Sanofi)
- Stefan Kutscheidt (Team Leader Quality Control, AGC Biologics, Heidelberg)
- Ellen Krautkrämer (Postdoc, Heidelberg University Hospital)
- Miguel Lamas (Clinical Research Associate, Argint International, Alicante, Spain)
- Benedikt Obermaier
- Xiao-Yu Pan (Senior Scientist and Manager, Boehringer Ingelheim, Shanghai)
- Virginia Pierini (EMBL)
- Francois Pujol (Becton Dickinson, Switzerland)
- Susanne Rauch (CureVac GmbH)
- Simin Rezaei
- Tatjana Reif (Associate Medical Operations, Biogen GmbH)
- Jochen Rudolph (Quality Manager, Miltenyi)
- Lopamudra Sadhu
- Jana Stastna
- Birthe Trautz (Scientific Coordinator, DKFZ, Heidelberg)
- Nikolaos Tsopoulidis (Postdoc, Massachusetts General Hospital, Harvard, Boston)
- Maud Trotard (ThermoFisher, Darmstadt)
- Ilka Wörz (Scientific Coordinator, Heidelberg University Hospital)
- Rebecka Wombacher (Postdoc, AbbVie, Ludwigshafen)
- Tobias Zierhut

