Group Breuhahn/Weiler
Transcriptional Regulation and Signaling Pathways in Hepatocytes and Liver Cancer Cells
With more than 700.000 new cases each year, hepatocellular carcinoma (HCC) is one of the leading causes of cancer-related death. Its risk factors are well-defined (e.g., chronic viral hepatitis, alcoholic and non-alcoholic steatohepatitis) and are detectable in up to 90% of all cases. However, the genetic heterogeneity of HCCs severely complicates the development of effective drugs, which is reflected by the current lack of efficient systemic therapies.
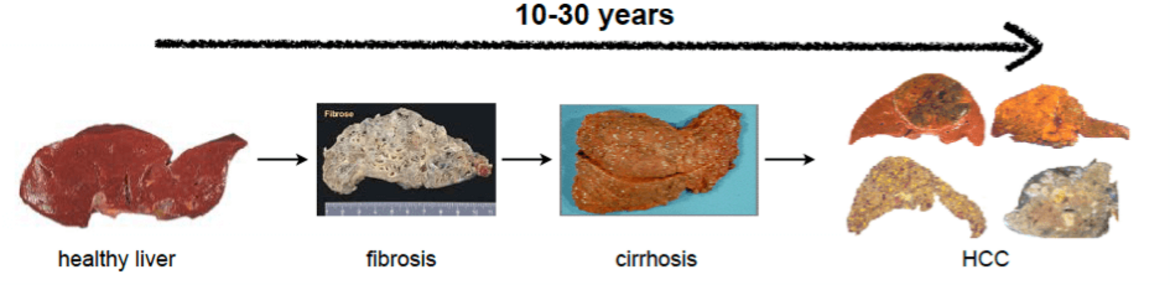
Liver cancer development is a multi-step process.
Different aetiologies lead to chronic hepatitis, which is characterized by continued cycles of hepatocyte death and tissue regeneration. Hepatic stellate cell activation and production of extracellular material mark progressed liver disease with fibrosis and cirrhosis. Mitotically active hepatocytes acquire somatic mutations and copy-number alterations that promote the formation of early HCC. Additional acquisition of epigenetic changes and alterations in molecular signaling pathways characterize HCC progression.
Transcriptional regulators (TRs) such as transcription factors or transcriptional co-activators, represent cellular bottlenecks in signal transduction. In carcinogenesis, several TRs and upstream regulators have been described as deregulated, leading to tumor cell proliferation and apoptosis resistance. Therefore, TRs are discussed as promising target structures for developing innovative therapeutic strategies. Our group focuses on the regulation and function of different TRs involved in regenerative processes as well as HCC development and progression. We want to understand how TRs support tumorigenesis and if they could serve as biomarkers for patient stratification. Our interdisciplinary approaches utilize techniques ranging from cell and molecular biology to bioinformatic methods. We investigate suitable in vitro and in vivo models and confirm findings in well-characterized human patient cohorts.
The Hippo signaling pathway is a sensor for cell density and negatively regulates its transcriptional coactivator yes-associated protein (YAP) and transcriptional coactivator with PDZ-binding motif (TAZ). In hepatocarcinogenesis, YAP represents an oncogene as the inactivation of Hippo pathway constituents (e.g., Mst1/2) or overexpression of YAP rapidly induces liver tumor formation in mice. We showed that YAP is overexpressed in a subgroup of human HCCs and that this factor supports tumor cell proliferation and invasion. Using cross-species comparison, the Notch receptor ligand Jagged-1 (Jag-1) was identified as a central downstream target gene in HCC and other gastrointestinal tumor types (Tschahargeneh et al., 2013 Gastroenterology). In addition, YAP induces a gene signature that characterizes HCC patients with chromosomal instability (CIN) (Weiler et al., 2017 Gastroenterology; Wei et al., 2019 Oncogene; Tóth et al., 2021 BMC Cancer).
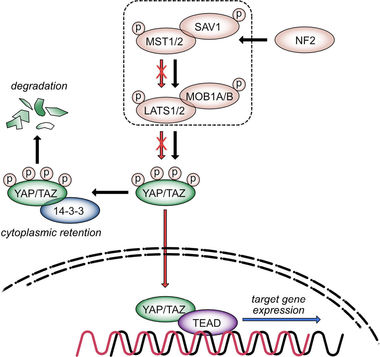
Scheme of Hippo signaling pathway. The Hippo kinase cassette consists of kinases (MST1/2, LATS1/2) and scaffold proteins (e.g., SAV1, NF2). Kinase-dependent YAP/TAZ phosphorylation is associated with their cytoplasmic retention and degradation (black arrows). Pathway inactivation leads to nuclear translocation of both transcriptional co-activators, where they interact with different TFs (e.g., TEAD1-4; red arrows). Binding of YAP/TF or TAZ/TF complexes in gene promoter regions stimulates the expression of factors controlling cell cycle and chromosomal segregation (blue arrow). Due to the negative impact of the kinase cassette on nuclear YAP/TAZ enrichment, the Hippo pathway is considered as tumor-suppressive, while its downstream effectors YAP/TAZ act as oncogenes in different model systems.
Our recent studies illustrate that YAP supports carcinogenesis via tumor cell-endogenous mechanisms and regulates heterologous communication networks. For example, overexpression of YAP in HCC cells controls the cellular secretome, directly impacting tumorous and non-tumorous cells (Thomann et al., 2020 Cancer Res; Marquard et al., 2020 Cell Commun Signal; Thomann et al., 2021 Liver Int, Liu et al. 2024 Cell Mol Life Sci).
In addition, we aim to implement systems biology methods to better understand how the Hippo pathway affects the biology in tumorous and non-tumorous liver cells. For example, we recently showed that the painkiller acetaminophen controls the dynamic shuttling process of YAP via nuclear phosphorylation processes. This was possible using sensitive live-cell imaging technology and mathematical pathway modeling (Wehling et al. 2022 eLife).
Another aspect of our research is how molecular 'upstream' mechanisms control the aberrant enrichment of YAP in hepatocytes and HCC cells. In this context, we demonstrated that target genes of the YAP paralogue TAZ are essential regulators of YAP activity in HCC cells (illustrated for integrin-αV; Weiler et al., 2020 Cancer Lett). In addition, we showed that cell junction proteins such as α‐catenin cooperate with Hippo/YAP signaling to control factors associated with cell migration and CIN (Tang et al., 2023 Cell Commun Signal).
Subproject of SME Weiler - Septins in cell physiology and carcinogenesis
Next to actin, microtubules and intermediate filaments, septins represent the fourth component of the cytoskeleton. We have recently shown that the YAP/TAZ target gene SEPTIN10 is upregulated in liver cancer conferring poor patient prognosis. SEPTIN10 serves as mechanotransducer by regulating actin and microtubule structures (Weiler et al., 2024 Cancer Lett). Our work studies the impact of septins on cellular behavior as well as their role during mechanotransduction.
Subprojekt of K Breuhahn - Long non-coding RNAs as cancer biomarkers
Long non-coding RNAs (lncRNAs) can influence liver tumorigenesis through various mechanisms. At the same time, they can also serve as biomarkers that indicate the activity of tumor-supporting signaling pathways or cellular processes. This project aims to define lncRNA signatures that provide insight into cellular processes in the tissue or serum of cancer patients. We hope to identify dysregulated oncogenes or signaling pathways early, even before imaging methods detect solid tumors.
Patent: Diagnostic methods and diagnostic assay comprising detection of lncRNAs (European patent application: 22210144.6 (filing: November 29, 2022; publication: June 5, 2024).
Publications
Wesener MC, Weiler SME, ... Breuhahn K. CRKL Enhances YAP Signaling through Binding and JNK/JUN Pathway Activation in Liver Cancer. Int J Mol Sci 2024; Aug 5;25(15):8549. (IF: 4.9)
Liu K, ... Weiler SME, ... Breuhahn K. Dynamic YAP expression in the non-parenchymal liver cell compartment controls heterologous cell communication. Cell Mol Life Sci 2024; 81: 115. (IF: 6.2)
Weiler SME, ... Breuhahn K. SEPTIN10-mediated crosstalk between cytoskeletal networks controls mechanotransduction and oncogenic YAP/TAZ signaling. Cancer Lett; 2024 Mar 1:584:216637. (IF: 9.1)
Tang Y, ... Weiler SME, ... Breuhahn K. α-catenin interaction with YAP/FoxM1/TEAD-induced CEP55 supports liver cancer cell migration. Cell Commun Signal 2023 Jun 28;21(1):162. (IF: 8.4)
Wehling L, ... Breuhahn K. Spatial modeling reveals nuclear phosphorylation and subcellular shuttling of YAP upon drug-induced liver injury. eLife. 2022 Oct 8;11:e78540. (IF: 7.7)
Tóth M, ... Weiler SME, ... Breuhahn K, Co-expression of YAP and TAZ associates with chromosomal instability in human cholangiocarcinoma. BMC Cancer. 2021 Oct 6;21(1):1079. (IF: 4.64)
Thomann S, Weiler SME, ... Breuhahn K. YAP-induced Ccl2 expression is associated with a switch in hepatic macrophage identity and vascular remodelling in liver cancer. Liver Int. 2021 Dec;41(12):3011-3023. (IF: 8.75)
Marquard S, ... Weiler SME, ... Breuhahn K. Yes-associated protein (YAP) induces a secretome phenotype and transcriptionally regulates plasminogen activator Inhibitor-1 (PAI-1) expression in hepatocarcinogenesis. Cell Commun Signal. 2020 Oct 23;18(1):166. (IF: 5.71)
Thomann S, Weiler SME, ... Breuhahn K. YAP Orchestrates Heterotypic Endothelial Cell Communication via HGF/c-MET Signaling in Liver Tumorigenesis. Cancer Res. 2020 Dec 15;80(24):5502-5514. (IF: 12.70)
Weiler SME, ... Breuhahn K. TAZ target gene ITGAV regulates invasion and feeds back positively on YAP and TAZ in liver cancer cells. Cancer Lett. 2020 Mar 31;473:164-175. (IF: 8.68)
Wei T, Weiler SME, ... Breuhahn K. YAP-dependent induction of UHMK1 supports nuclear enrichment of the oncogene MYBL2 and proliferation in liver cancer cells. Oncogene. 2019 Jul;38(27):5541-5550. (IF: 6.6)
Wan S, ... Weiler SME, ... Breuhahn K. Cytoplasmic localization of the cell polarity factor scribble supports liver tumor formation and tumor cell invasiveness. Hepatology. 2018 May;67(5):1842-1856. (IF: 15.0)
Weiler SME, ... Breuhahn K. Induction of Chromosome Instability by Activation of Yes-Associated Protein and Forkhead Box M1 in Liver Cancer. Gastroenterology. 2017Jun;152(8):2037-2051. (IF: 20.8)
Tschaharganeh DF, ... Breuhahn K. Yes-Associated Protein Up-regulates Jagged-1 and Activates the NOTCH Pathway in Human Hepatocellular Carcinoma. Gastroenterology. 2013 Jun;144(7):1530-1542. (IF: 13.9)
Straßburger K, ... Breuhahn K, Teleman AA. Insulin/IGF signaling drives cell proliferation in part via Yorkie/YAP. Dev Biol. 2012 Jul 15;367(2):187-96. (IF: 3.2)
Breuhahn K, Schirmacher P. A cellular view of Nf2 in liver homeostasis and tumorigenesis. Dev Cell. 2010 Sep 14;19(3):363-4. (IF: 9.3)
Current group members
Dr. rer. nat. Sofia ME Weiler
Dr. rer. nat. Amruta Damle-Vartak
Fabiola Pedrini
Michaela Bissinger
Jennifer Schmitt
Elias Klossok
Solomiia Savchenko
Bianca Metze (laboratory support)
Currently not in the lab
Active collaborations
Prof. Arndt Vogel (MHH, Medizinische Hochschule Hannover)
PD Dr. Anna Saborowski (MHH, Medizinische Hochschule Hannover)
Prof. Heike Bantel (MHH, Medizinische Hochschule Hannover)
PD Dr. Katja Breitkopf-Heinlein (UMM, University Hospital Mannheim)
Prof. Jan Hengstler (IfADo, Dortmund)
Dr. Nachiket Vartak (IfADo, Dortmund)
Prof. Ahmed Abdelaziz Fahmy (NewGiza University, Cairo, Egypt)
Prof. Stephan Singer (Institute of Pathology, University Hospital Tübingen)
Prof. Mathias Heikenwälder (Deutsches Krebsforschungszentrum, DKFZ)
Our work is currently supported by
Deutsche Forschungsgemeinschaft (DFG)
Deutsche Krebshilfe
Physician Scientist Programm of the Medical Faculty Heidelberg (Dr. S. Weiler)
Open positions
We continuously welcome applications from persons with external funding sources.
For ambitious medical students we offer experimental projects that require two semesters full-time commitment.
We offer the possibility of performing the experimental part of a Master's Thesis in our group. Supervision by experienced scientists or PhD students is ensured.
Former group members
Stefania Bonusi (Master student)
Patrizia Birner (technician)
Tobias Klessinger (medical student)
Camilla Maggi (Master student)
Dr. Lena Thiess (PhD student)
Dr. Yingyue Tang (medical student)
Dr. Klára Schulc (guest scientist)
Dr. Fabian Rose (PhD student)
Dr. Lilija Wehling (PhD student, scientist)
Fabian von Bubnoff (medical student)
Marie Wesener (medical student)
Dr. Paula Fernández-Palanca (guest scientist)
Dr. Kaijing Liu (medical student)
Dr. Nada El-Ekiaby (guest scientist)
Dr. Dr. Stefan Thomann (PhD student, scientist)
Dr. Teng Wei (Dr. sc. hum. student)
Dr. Simone Marquard (medical student)
Dr. Margarita González-Vallinas Garrachón (scientist)
Dr. Federico Pinna (scientist)
Dr. Jana Samarin (diploma student, scientist)
Dr. Mona Malz (PhD student, scientist)
Dr. Sabrina Schmitt (scientist)
Dr. Maria Knaub (PhD student)
Dr. Shan Wan (PhD student)
Dr. Benedikt Müller (PhD student)
Dr. Antje Brauckhoff (PhD student)
Dr. Tanja Nussbaum (PhD student)
Dr. Stephanie Schnickman (PhD student)
Dr. Sebastian Vreden (PhD student)
Dr. Charlotte Jankovitz (medical student)
Dr. Teresa Lutz (medical student)
Dr. Philipp Latzko (medical student)
Dr. Michael Bovet (medical student)
Dr. Pia Moinzadeh (medical student)
Anne-Sophie Meyer (physician)
Dr. Christian Rupp (physician)
Prof. Darjus Tschaharganeh (physician)
Prof. Stephan Singer (physician)
Vera Riehmer (diploma student)
Yelena Burda (diploma student)
Ute Müller (technician)
Petra Hubbe (technician)
Martina Keith (technician)
Our nationalities

Contact
Prof. Dr. Kai Breuhahn
Dr. Sofia ME Weiler
University Hospital Heidelberg
Institute of Pathology
Im Neuenheimer Feld 224
69120 Heidelberg
+49-6221-56-4675
kai.breuhahn(at)med.uni-heidelberg.de
sofia.weiler(at)med.uni-heidelberg.de
Last update: September 2024