- Section Nephrogenetics (AG Simons)
- Seniorprofessorship Prof. Rappold
- Molecular Neurogenetics (AG Berkel)
- Cardiogenetics (AG Hoffmann)
- Neurogastrogenetics
- Transcriptional regulation in developmental disorders (AG Laugsch)
- Genetics of neurodevelopmental disorders (AG Schaaf)
- Mouse models for neurodevelopmental disorders
- Research in our Diagnostic Laboratories
- Publications
Transcriptional Regulation in Developmental Disorders
POSTDOCTORAL POSITION IN DEVELOPMENTAL BIOLOGY AND DISEASE MODELING WITH COMBINED WET (70%) AND DRY LAB (30%) WORK (D/F/M)
We are looking for an enthusiastic postdoc to join our research group at the Institute of Human Genetics of the University Heidelberg (Germany) to study the pathophysiology of Cornelia de Lange Syndrome (CdLS).
Following generation of CRISPR/Cas9 engineered human induced pluripotent stem cells (hiPSC) with CdLS mutations, the successful candidate will characterize the hiPSC as well as hiPSC-derived neural crest cells (NCC) by HiChIP-seq, RNA-seq, and ATAC-seq. Integrative analysis of these data should reveal the regulatory networks controlled by cohesin in hiPSC and hiPSC-derived NCC
Our laboratory explores early human development using hiPSC and their differentiation to understand gene regulation by non-coding DNA elements and the 3D chromosomal architecture in health and disease. We incorporate cutting-edge methods from various disciplines (cell biology, genomics, gene editing, bioinformatics, network biology) to provide a holistic picture of the most fundamental developmental processes.
Composed of international and multi-disciplinary scientists, our group creates a unique and inspiring environment and supports individual career development. For more information visit:
Project relevant literature
doi: 10.1016/j.stem.2019.03.004.
doi: 10.1177/00220345211038203.
doi: 10.1371/journal.pone.0065149.
Your profile:
- Ph.D.in bioscience or equivalent degree with substantial experience in stem cell biology and basic bioinformatics.
- Cell culture experience and standard molecular biological techniques such as cloning, microscopy, Western blot, immunofluorescence, qPCR is a must.
- Experience in at least one of the following techniques is required: ChIP-seq, ATAC-seq, Hi-C, or 4C-seq is required.
- Experience in hiPSC culture, CRISPR/Cas9 targeting and their differentiation is a huge advantage
- Ability to analyze own sequencing data as well as strong interest in Python, R, shell scripting and working with NGS data.
- Curiosity-driven researcher, passionate about science, who can work both independently and as part of our team.
- Strong written and verbal communication skills in English.
Duration:
Twenty months with a potential option to extend, starting as soon as possible.
Contact and Application:
The application must include your motivation, a brief statement of your scientific interests including your contribution to the projects you were working on and how you can contribute to the cohesin project. In addition, contact details from three references, curriculum vitae, separated publication list, and relevant certificates.
Application deadline: 31.05.2025
Please forward your complete application in a single pdf document with the filename: Postdoc_ Cohesin25_ first name_ second name (no other format will be accepted) by e-mail to magdalena.laugsch(at)uni-heidelberg.de
WHO?

WHAT?
We explore the relationship between craniofacial and brain development and their intimate interactions in health and disease.
WHY?
In vertebrates, the head forms from a transient and multipotent population of neural crest cells that appears after the embryonic neural tube closes. From this point onward, head and brain formation are intertwined throughout development. The head protects the developing brain and facilitates its functional integration, and many neurodevelopmental disorders also include craniofacial malformations. Our research focuses on developmental genes that need a specific and dynamic regulation e.g. by enhancers. That regulation ensures establishing their precise expression patterns during development, which might have pathological consequences when being disrupted. In this context, our investigations of the similarities and differences between human neural crest cells and neuronal cells will provide valuable insights into the causes of neurodevelopmental conditions.
HOW?
A great breakthrough in the field of human disease modeling was the possibility to obtain human stem cells by reprogramming adult somatic cells. Since then, these so-called human induced pluripotent stem cells (hiPSC) have been generated from patient donors and then differentiated into disease-relevant cell types that can be used to expand our knowledge about the molecular basis of disease. The power of disease modeling has further expanded with the development of CRISPR/Cas9 technology, which simplifies introducing or correcting mutations in cellular DNA.
Leveraging our expertise in hiPSC culture, genome editing (CRISPR/Cas9), and their directed differentiation (neural and neural crest), we have been recapitulating early human embryogenesis. To analyze that process, we have been combining epigenomic (ChIP-seq, 4C-seq, Hi-ChIP-seq) and transcriptomic data (RNA-seq) with advanced computational approaches. Our genomewide data should provide an excellent foundation for innovative studies on the intimate interactions and relationships between craniofacial and brain development.
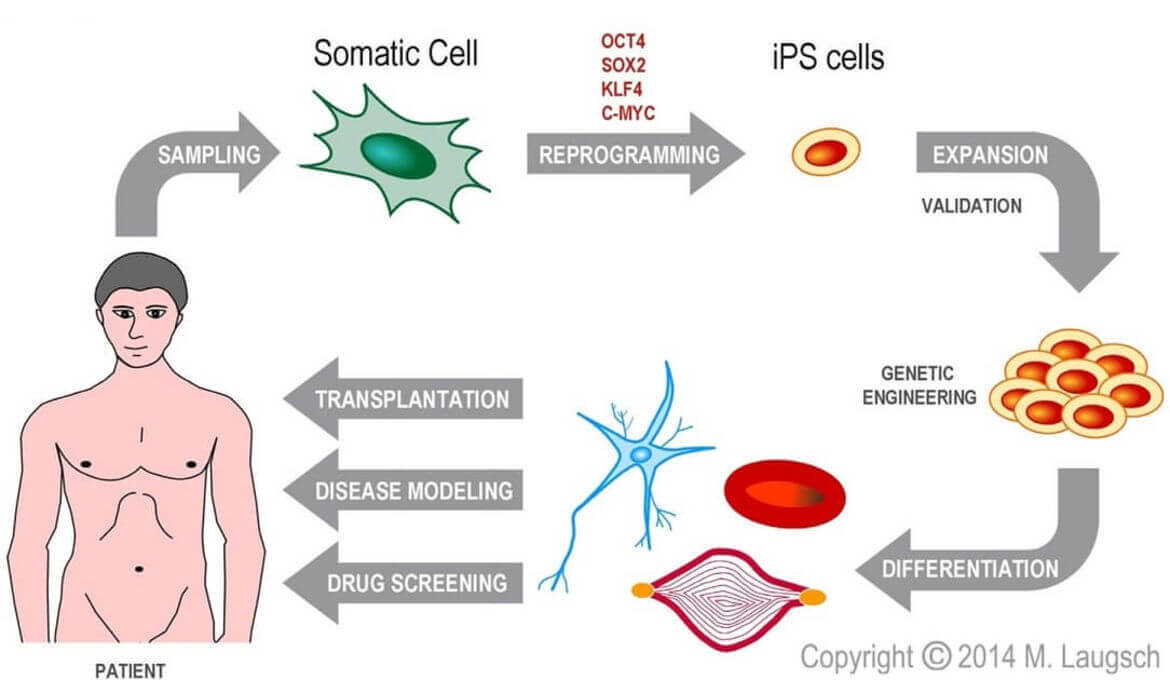
Figure 1: Scheme of generation and possible applications of hiPSC. Human cells of any origin can be reprogramed into hiPSC by transient overexpression of a combination of certain transcription factors. Due to their renewing potential, hiPSC can be expanded and engineered. Finally, hiPSC can be differentiated into the desired cell type and applied for disease modelling, drug screening or transplantation.
Join the lab
YOU are a curiosity-driven researcher, passionated for science, have good written and verbal communication skills in English, can work both independently and as part of our team.
WE offer you a creative and inspiring environment of highly enthusiastic and motivated scientists. You will find here an international and multi-disciplinary group and a friendly working atmosphere that supports individual career development. Our team and our outstanding national and international collaborations apply cutting-edge approaches to investigate the craniofacial and brain development using hiPSC models.
MASTER AND BSC. STUDENTS, INTERN students
Talented students with a strong interest in stem cell, molecular biology, and bioinformatics are welcome in our laboratory. However, the opportunities strongly depend on our ongoing scientific questions and capacities.
OPEN PHD AND POSTDOC POSITIONS
Currently, there are no open positions available, but excellent candidates are always encouraged to contact Magdalena Laugsch.
SELECTED RECENT PUBLICATIONS
Overarching control of autophagy and DNA damage response by CHD6 revealed by modeling a rare human pathology.
Y. Kargapolova, R. Rehimi, H. Kayserili, J. Brühl, K. Sofiadis, A. Zirkel, S. Palikyras, A. Mizi, Y. Li, G. Yigit, et al.
Nat Commun. 2021 May 21;12(1). doi: 10.1038/s41467-021-23327-1.
Dosage analysis of the 7q11.23 Williams region identifies BAZ1B as a major human gene patterning the modern human face and underlying self-domestication.
M. Zanella, A. Vitriolo, A. Andirko, P. T. Martins, S. Sturm, T. O’Rourke, M. Laugsch, N. Malerba, A. Skaros, S. Trattaro, et al.
Sci. Adv. 2019 Dec 6;5(12). doi: 10.1126/sciadv.aaw7908.
mTOR and autophagy pathways are dysregulated in murine and human models of Schaaf-Yang syndrome.
E. Crutcher, R. Pal, F. Naini, P. Zhang, M. Laugsch, J. Kim, A. Bajic, C. P. Schaaf.
Sci Rep. 2019 Nov 4;9(1). doi: 10.1038/s41598-019-52287-2.
Modeling the Pathological Long-Range Regulatory Effects of Human Structural Variation with Patient-Specific hiPSCs.
M. Laugsch*, M. Bartusel*, R. Rehimi, H. Alirzayeva, A. Karaolidou, G. Crispatzu, P. Zentis, M. Nikolic, T. Bleckwehl, P. Kolovos, et al.
Cell Stem Cell. 2019 May;24(5):736-752.e12. doi: 10.1016/j.stem.2019.03.004.
Functional Restoration of gp91phox-Oxidase Activity by BAC Transgenesis and Gene Targeting in X-linked Chronic Granulomatous Disease iPSCs.
M. Laugsch*, M. Rostovskaya*, S. Velychko, C. Richter, A. Zimmer, B. Klink, E. Schröck, M. Haase, K. Neumann, S. Thieme, et al.
Molecular Therapy. 2016 April;24(4):812-822. doi: 10.1038/mt.2015.154.
* shared authorship
Group members
Group Leader Dr. rer. nat. Magdalena Laugsch
After her diploma studies in biology (University of Braunschweig and University of Cologne), she obtained her Ph.D. degree at the Institute of Pharmacology and Toxicology (University of Magdeburg). For her following career, scientific projects with Prof. Rolf Jessberger (Institute of Biochemistry, Dresden) and Dr. Alvaro Rada-Iglesias (CMMC, Cologne) were crucial. In Prof. Schaaf's lab (2019), she got the great opportunity to study neurodevelopmental disorders related to the neural crest (Institute of Human Genetics, Heidelberg). In 2020, Magdalena became a group leader. Since then, she is expanding her research in this area.
Contact
+49 6221 56-39128
Magdalena.Laugsch(at)uni-heidelberg.de
Aylin Camgöz, PhD (collaboration with Dr. Marcel Kool) Aylin received her doctoral degree from Frank Buchholz lab at the department of Medical Systems Biology at the University Cancer Center, Dresden. She continues her scientific journey as a postdoctoral researcher in the Pediatric Neuro-oncology department at DFKZ. She currently investigates the function of epigenetic regulators in the initiation and maintenance of pediatric ependymoma.
Ayat Ahmed, MSc
Ayat received her Master of Molecular Biology of the Cell at the Faculty of Science and Technology in Milan, Italy and accomplishes her PhD in our lab. Her research focuses on investigating the regulatory landscape of the transcription factor NR2F1 in the neural crest. Besides, she aims to determine the regulatory network controlled by NR2F1 in neural crest and neurodevelopmental disorder.
Email: Ayat.Ahmed@med.uni-heidelberg.de
Ragini Gabhrani, intern
Ragini is pursuing her master’s in Molecular Biotechnology from the Anhalt University of Applied Sciences. She is joining the lab from November, 2022 to undertake an internship followed by her master’s thesis. Her main focus is on the characterization of neural crest and neuronal cells derived from hiPSC of BBSOAS patients.
Stella Bähr, intern
Stella is currently pursuing her Masters Degree in Molecular Biotechnology at the University of Heidelberg, with a focus on Bioinformatics. She joined the lab to accomplice her intern and will analyze publically available data by computational tools to expand our understanding of the NR2F1 transcription factor.
Susanne Theiß, Technician
Susanne is working on genome organization (4C and Hi-C) and identifying genome-wide DNA binding sites for different transcription factors and histone modifications (ChIP-seq). She supports us in determining changes in regulatory landscapes and networks in various neurodevelopmental disorders. In addition, Susanne is an expert in the field of CRISPR/Cas9 mediated genome editing.
Email: Susanne.Theiss@uni-heidelberg.de
Ruiyuan Zheng, Postdoc
Having received doctoral education in the University of Cologne, Ruiyuan Zheng is now joining the group to study the impact of cohesin on human head and face development as a postdoc. She focuses on bioinformatical analysis of the experimental data generated in during neural crest cell differentiation.
Büşranur Çeltik, intern
Büşra recently got her undergraduate degree in Genetics and Bioengineering at Izmir University of Economics in Turkey and joined the lab for an internship. She aims to examine the role of cohesin by knocking down one of its subunits in hiPSC cells.
Jannis Bücking, medical student
As a medical student at the University of Heidelberg, Jannis is completing his medical dissertation by studying the neurodevelopmental disorders Prader-Willi syndrome and Schaaf-Yang syndrome. In doing so, he applies the CRISPR/Ca9 technique to knock out genes relevant to the development of the diseases and differentiates iPSCs into neurons.
Email: Jannis.Buecking@stud.uni-heidelberg.de
Freya Herrmann-Sim, intern
Freya is an undergraduate student of biosciences at the University of Heidelberg and joined the lab in September 2022 for an internship. By applying the CRISPR/Cas9 technique, she generates hiPSC lines with deletions of MAGEL2 to gain further understanding of the neurodevelopmental disorders Prader-Willi and Schaaf-Yang syndrome.
Tobias Beschauner, BSc
Tobias is a master's student of Biochemistry at the University of Heidelberg. During his internship he investigates the role of MAGEL2 in the Schaaf-Yang and Prader-Willi syndrome in cortical neurons derived from healthy and CRISPR/Cas9 engineered hiPSC.
Aaron Sievers, MSc
Aaron is a biophysicist working on in silico studies of DNA-sequence-dependent physiochemical properties of chromatin. He is responsible for bioinformatic analysis.
Email: Aaron.Sievers@med.uni-heidelberg.de
Cao Ding, MSc (collaboration with Dr. Christina Eichstaedt)
Cao is a student in the doctor of medicine program. He investigates the functional effect of genetic variants of pulmonary arterial hypertension patients.
Email: Cao.Ding@med.uni-heidelberg.de
Alumni
Aaron Sievers, MSc (biophysicist, was responsible for bioinformatic analysis)
Feven Berhanne (intern and bachelor thesis 2022, Applied Biology at the University of Applied Sciences Bonn-Rhein-Sieg in Rheinbach)
Luis Castillo technician (2021/2022, supported us as a technical assistant in the laboratory)
Tamara Schmidt, BSc, intern (Winter/Summer 2022, Biochemistry Master studies, University of Heidelberg)
Leonie Schumacher, intern (Spring/Summer 2022, Villingen-Schwenningen University Gemany)
Mara Rudigier, intern (bachelor thesis 2022, Villingen-Schwenningen University Gemany)
Melanie Spanjaard, medical student (medical dissertation 2022, University of Heidelberg)
Kristina Krank, BSc (intern, June-November 2021, Biochemistry Master studies, University of Heidelberg
Arya Lal Erkilinçoğlu (intern, Summer 2020, student of Yeditepe University, Istanbul, Turkey)
Sarah Cluff, MSc (master thesis, July 2021, Molecular Cellular Biology, University of Heidelberg)
Marc Thomas, BSc (intern, Spring 2021, Biochemistry Master studies, University of Heidelberg)
Alexander Wirth, BSc (intern, 2020/2021, Biochemistry Master studies in University of Heidelberg)
Rocio Olmos Romero, MSc (master thesis, December 2020, Translational Medical Research, University of Heidelberg)
- Section Nephrogenetics (AG Simons)
- Seniorprofessorship Prof. Rappold
- Molecular Neurogenetics (AG Berkel)
- Cardiogenetics (AG Hoffmann)
- Neurogastrogenetics
- Transcriptional regulation in developmental disorders (AG Laugsch)
- Genetics of neurodevelopmental disorders (AG Schaaf)
- Mouse models for neurodevelopmental disorders
- Research in our Diagnostic Laboratories
- Publications
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/8/2/csm_20131204_Beratung_035_a396c6c6e5.jpg)
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/a/0/csm_20170627_PflegeOrtho_001_fb912471fa.jpg)
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/f/c/csm_20170215_LaborOMZ_155_c0169c0898.jpg)
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/2/c/csm_20180523_Labor_139_6ebb9a0a1b.jpg)
