THE NCOUNTER CORE FACILITY
The nCounter Core Facility was established in 2011 and is part of the CellNetworks Core Technology Platform at Heidelberg University and the MULTI-SPACE platform of the Health & Life Science Alliance.
The facility offers state-of-the-art methods for mRNA and miRNA expression profiling (nCounter SPRINT Profiler), as well as Digital Spatial Profiling at the transcript level (GeoMx Digital Spatial Profiler) and protein level (GeoMx Digital Spatial Profiler, Lunaphore COMET).
In addition, the facility provides various analytical methods for quality control of RNA and DNA samples.
In the DFG’s research infrastructure portal "RIsources", the nCounter Core Facility is listed under RI_00563 (nCounter SPRINT Profiler, GeoMX Digital Spatial Profiler) and RI_00613 (Lunaphore COMET).
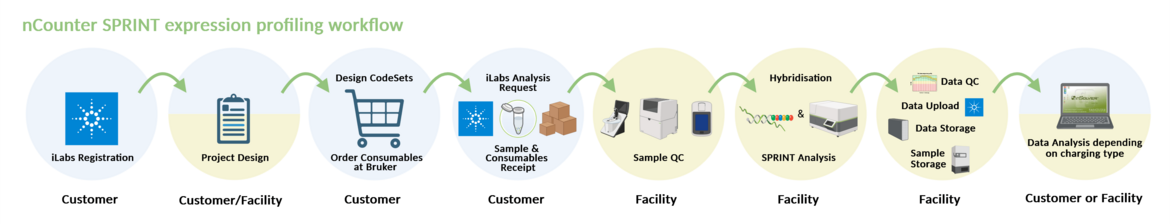
THE NCOUNTER SPRINT PROFILER
The nCounter SPRINT Profiler from Burker Spatial Biology is suitable for digital expression analysis of up to 800 mRNAs or miRNAs as well as CNV analysis at DNA level.
For multiplex analysis, the device uses fluorescently labelled reporter probes, which are highly resistant to poor RNA quality due to their length of approx. 100 bases and are therefore ideal for samples that are difficult to analyse, such as FFPE(formalin-fixed, paraffin-embedded) samples.
The application of a unique coding technology enables the direct counting of single RNA molecules at all levels of biological expression, with a sensitivity and specificity comparable to Real Time PCR (RT PCR). In addition, only very small amounts of RNA (25-150ng) or DNA (150ng) are required for the analysis. Reverse transcription of the RNA is not required. The SPRINT Profiler is a perfect system for the validation of microarrays, the analysis of signalling pathways, the expression analysis of defined gene lists (individual or predefined) as well as for the validation of biomarkers.
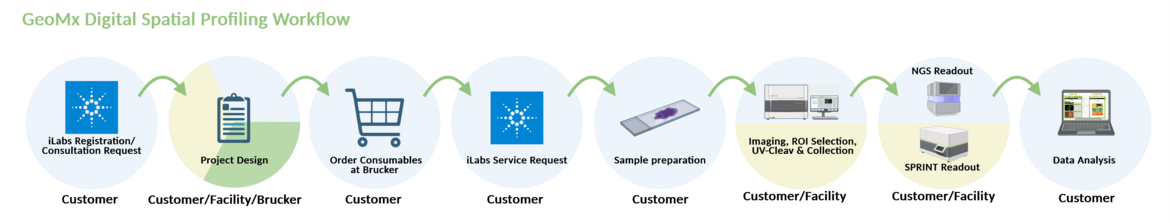
THE GEOMX DIGITAL SPATIAL PROFILER
For multicellular expression profiling at the transcript and protein level, the nCounter Core Facility offers the GeoMx Digital Spatial Profiler from Burker Spatial Biology, which uses light-cleavable oligonucleotide barcodes bound to antibodies or RNA probes that enable almost unlimited multiplexing capability (~20 000 RNAs, ≥100 proteins). In addition, the device allows highly complex spatial resolution and quantification of proteins and RNA (Whole Transcriptome Read-out).
LUNAPHORE COMET PLATFORM
The Lunaphore COMET platform(https://lunaphore.com/products/comet) enables the simultaneous visualisation of up to 40 antibodies on one to four tissue samples. The automated workflow and a precise microfluidics system ensure high reproducibility. In addition to proteins, specific RNA samples can also be stained.
The tissue samples (FFPE or Fresh Frozen) are directly or indirectly stained and imaged. The background is automatically subtracted. The generated images are saved in tiff format and can therefore be analysed with common programs.
Workflow Lunaphore COMET platform:
After registering with iLabs and requesting a "Lunaphore COMET Consultation request" in iLabs, the project and the experiments are discussed. After completing the "Lunaphore COMET sample run" request in iLabs and sending all required materials and samples, test runs are carried out first and then the experiments themselves after further discussion. The data can then be collected or downloaded as agreed.
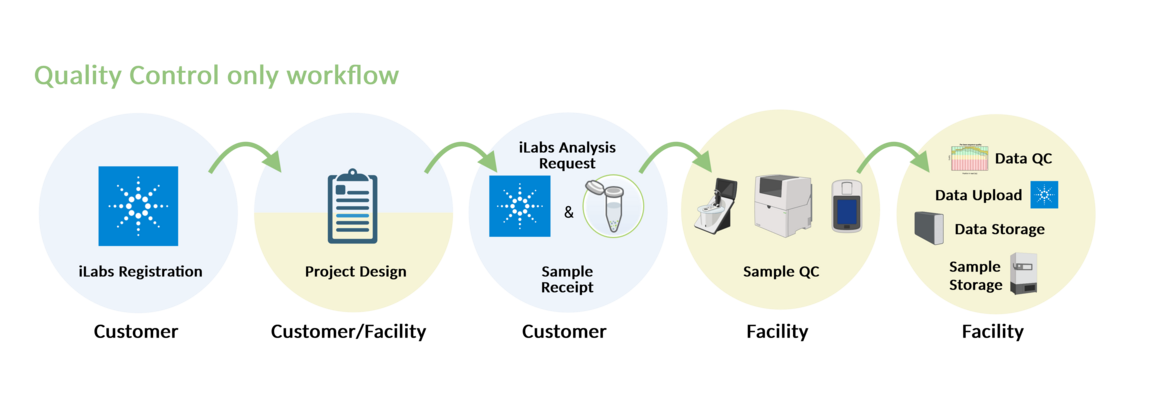
QUALITY CONTROL
For pure quality control analyses, we use the Agilent TapeStation and the Qubit system from Thermo Fisher Scientific.
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/8/2/csm_20131204_Beratung_035_a396c6c6e5.jpg)
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/a/0/csm_20170627_PflegeOrtho_001_fb912471fa.jpg)
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/f/c/csm_20170215_LaborOMZ_155_c0169c0898.jpg)
![[Translate to English:] [Translate to English:]](/fileadmin/_processed_/2/c/csm_20180523_Labor_139_6ebb9a0a1b.jpg)





