AG Raffel - Emmy Noether Programm
“Targeting leukemia stem cell vulnerabilities”
In our research lab, we aim to identify targetable vulnerabilities in leukemic stem cells (LSCs). By dissecting their fundamental biology, we develop novel diagnostic tools and therapeutic strategies to identify and eradicate LSCs. Our ultimate goal is to improve clinical outcomes of patients suffering from acute leukemias.
We mainly focus on acute myeloid leukemia (AML), which is an aggressive hematological malignancy characterized by the accumulation of clonal myeloid blasts unable to differentiate into mature blood cells. Chemotherapy induces remission in the majority of patients, but relapse rates are very high and lead to poor clinical outcomes. Relapse and refractoriness are caused by chemotherapy-resistant leukemic stem cells (LSCs) and in order to improve patient survival, it is therefore essential to eradicate LSCs. We characterize LSC populations at multiple layers, including proteome, transcriptome, genome, metabolome, epigenome, and cellular function. Due to the plasticity of LSC phenotypes, the identification of surface markers to target LSC by immunotherapeutic approaches such as bispecific antibodies or CAR-T cells, remains a challenge. Alternative strategies to interfere with rather fundamental aspects of tumor cell biology, such as metabolism or epigenetic alterations, may be more broadly effective despite inter- and intra-patient heterogeneity of LSC.
We also develop strategies to characterize LSCs at the single cell level. We simultaneously analyze metabolism, gene expression, mutational landscapes, and cellular function of thousands of individual leukemic cells. Using this approach, we are able to dissect clonal and hierarchical heterogeneity of leukemic cells, and, most importantly, identify and functionally test rare LSCs in a complex mixture of healthy and diseased cells within the human bone marrow of leukemia patients.
More recently, we have broadened the focus and use our technologies to study other hematologic diseases, including acute lymphoblastic leukemia (ALL).
The lab is funded by the Emmy Noether Programme of the German Research Foundation (DFG) and within the Junior research alliance “LeukoSyStem”) of the German Federal Ministry of Education and Research.
Current projects
Targeting energy, amino acid, and lipid metabolism in LSCs
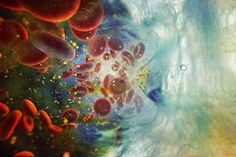
To study fundamental biologic properties of LSCs, we characterized patient-derived LSC and blast populations as well as healthy blood stem cells by quantitative multiplex proteomic analyses and RNA-Sequencing (Raffel et al., Nature 2017; Raffel et al., Blood 2020). As there is no universal phenotypic definition of LSCs, we analyzed their functional capacity to re-initiate the leukemia after xenotransplantation into immuno-compromised mice (Figure 1A). This in vivo assay remains the gold standard for the detection of LSCs. Our data underscores the difficulties in identifying reliable LSC markers that can be exploited for diagnostics or antibody-based therapies. The proteome is particularly powerful in evaluating metabolic properties (Figure 1B). We identified several metabolic pathways in LSCs, such as oxidative phosphorylation, phospholipid and fatty acid metabolism as well as degradation of branched chain amino acids (BCAA). BCAA degradation by BCAT1 plays a critical role for alpha-ketoglutarate (αKG) homeostasis. αKG is an essential co-substrate for cellular dioxygenases, which control a broad range of cellular functions. Suppression of αKG-dependent enzymes constitutes a common feature of AML and, next to mutations in IDH and TET2, we identified the BCAA–BCAT1–αKG axis as an alternative, non-genetic mechanism in LSCs to control the activity of these enzymes (Figure 1C).
We are currently exploring specific vulnerabilities in energy, amino acid, and lipid metabolism in LSCs. We apply state-of-the-art technologies in primary AML specimen, including targeted and untargeted metabolomics, lipidomics, proteomics, single-cell multiomics, CRISPR gene knockouts, lentivirus-mediated knockdown or overexpression, stromal co-cultures, and in vivo experiments. We have access to large clinical cohorts and the biobank of our clinic.
Dissecting metabolism, gene expression, clonality and function of LSCs at single-cell resolution
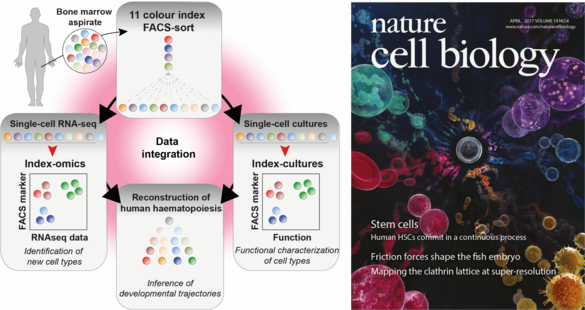
Figure 2: Combined transcriptomic and functional single cell approach to study the human healthy and malignant hematopoietic system.
Using single cell sequencing technologies, heterogeneity can be resolved from complex mixtures of cells. As LSCs require a functional assay, we developed a platform for simultaneous single-cell transcriptomic and single-cell culture analyses (Velten*, Haas*, Raffel* et al., Nature Cell Biology 2017). By index-FACS sorting and mathematical modeling, gene expression analysis (which requires lysis of cells) and single cell culture assays (which require living cells) can be quantitatively linked at single cell resolution (Figure 2).
In ongoing experiments, we unite our biomedical and technological expertise to study fundamental aspects of metabolism, gene expression, clonal heterogeneity, and stem cell function of LSCs at the single cell level. We combine flow cytometric analysis of metabolic properties and surface marker expression with transcriptomic, mutational, and functional data at the single cell level. The application of index-FACS sort strategies and mathematical modeling enables us to integrate all these properties simultaneously in single cells. This will help to de-convolute heterogeneity, detect, and define putative markers for true LSCs, and identify targets to interfere with specific metabolic pathways characteristic for LSCs.
Furthermore, together with our close collaborators Simon Haas (Charite, Berlin) and Lars Velten (CRG, Barcelona) we perform comprehensive single cell analyses from large cohorts of AML patient samples of all different AML subtypes applying CITE-seq technologies and spectral flow cytometry.
More recently, we broadened our focus and apply the above-mentioned technologies to other hematologic diseases, including acute lymphoblastic leukemia (ALL).
Making leukemia stem cells vulnerable to immunotherapy
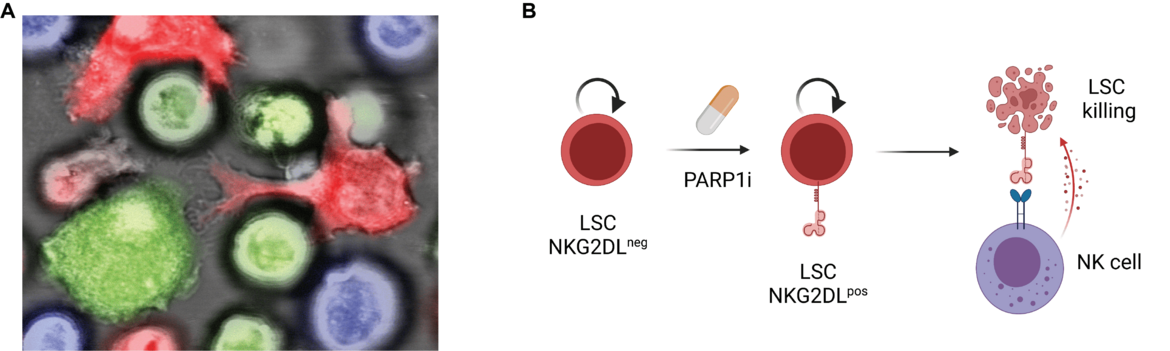
LSC biology is driven by genetic, epigenetic and metabolic alterations that promote LSC function. Besides these cell-intrinsic properties, LSCs are likely to escape surveillance of the immune system in order to promote tumorigenesis. Immune-checkpoint blockade has revolutionized therapeutic concepts in solid tumors. However, these T cell related checkpoints only play a minor role in AML. Therefore, we analyzed the NKG2D/NKG2DL-axis (natural killer group 2 member D and its ligands), which is a critical mediator of anti-tumor immunity by NK cells (Paczulla*, Rothfelder*, Raffel* et al., Nature 2019). Functional LSCs are characterized by the absence of NKG2D-ligands, which renders them “invisible” for NK cells. Polyclonal NK cells (pNKC) can kill AML blasts but not LSCs (Figure 3A). We identify PARP1 as critical regulator of NKG2DL expression in AML and chemical PARP1 inhibition induces NKG2DL expression on the surface of LSCs, which subsequently can be killed by pNKC transfer (Figure 3B). Treatment of mice transplanted with human AML cells with PARP1 inhibitors, followed by administration of pNKC, inhibited long-term in vivo leukemogenesis. These observations are of particular interest because PARP1 inhibitors are widely used for other indications in clinical practice. Based on our results, we have now initiated a clinical trial at our clinic with the combination treatment of PARP1 inhibition followed by pNKC transfusion aiming to eradicate LSC that otherwise evade the control of the immune system (NAKIP-AML, NCT05319249 (link: www.clinicaltrials.gov/ct2/show/NCT05319249), LKP: Prof. C. Müller-Tidow).
In ongoing projects, we aim to render LSCs even more susceptible to immunotherapy by optimizing pharmacological induction of NKG2DL expression, identification of co-upregulated surface antigens, and targeting both NKG2DL and the identified antigens using a combined CAR-NK/CAR-T cell product.
Team
Arbeitsgruppenleiter
Labormanagement
Wiss. Mitarbeiter/-innen
MD students
-

Lukas Franz
-

Selina Neunhäuser
-

Fabian Schulte
Technician
Postdoctoral Researcher
Collaboration Partners
Dr. Simon Haas, Charité, Berlin
Dr. Lars Velten, CRG, Barcelona
Dr. Laleh Haghverdi, Charité, Berlin
Dr. Gernot Poschet, Centre for Organismal Studies, Metabolomics Core Technology Platform (MCTP), Heidelberg University
Prof. Andreas Trumpp, German Cancer Research Center (DKFZ) and Heidelberg Institute for Stem Cell Technologies and Experimental Medicine (HI-STEM)
Dr. Stefano Casola, IFOM, the FIRC Institute of Molecular Oncology, Milan, Italy
Vacancies
We are recruiting highly motivated PhD and MD students. Please contact simon.raffel(at)med.uni-heidelberg.de
Press releases
Manfred Fuchs-Preis der Heidelberger Akademie der Wissenschaften für PD Dr. Simon Raffel
Maximilian Mönnig erhält José Carreras-DGHO-Promotionsstipendium
2,45 Millionen Euro zur Erforschung von Ursachen und Therapie bei Leukämie
Vier Heidelberger Doktoranden erhalten Stipendien für ihre Leukämie-Forschung
Forschung: Blutkrebs-Stammzellen auf der Spur
Double success for Heidelberg stem cell researchers
Misregulated protein breakdown promotes leukemias and brain cancer
Sibling Rivalry: how therapies for B cell lymphomas may give rare tumor cells an edge
Human blood cells develop in a continuous process instead of a stepwise one